A molecular phylogeny of tortoises (Testudines: Testudinidae) based on mitochondrial and nuclear genes
|
|
|
- Caroline Casey
- 6 years ago
- Views:
Transcription
1 Molecular Phylogenetics and Evolution 40 (2006) A molecular phylogeny of tortoises (Testudines: Testudinidae) based on mitochondrial and nuclear genes Minh Le a,b,, Christopher J. Raxworthy a, William P. McCord c, Lisa Mertz a a Department of Herpetology, Division of Vertebrate Zoology, American Museum of Natural History, Central Park West at 79th Street, New York, NY 10024, USA b Department of Ecology, Evolution, and Environmental Biology (E3B), Columbia University, 2960 Broadway, New York, NY 10027, USA c East Fishkill Animal Hospital, 455 Route 82, Hopewell Junction, New York 12533, USA Received 9 October 2005; revised 1 March 2006; accepted 2 March 2006 Available online 5 May 2006 Abstract Although tortoises of the family Testudinidae represent a familiar and widely distributed group of turtles, their phylogenetic relationships have remained contentious. In this study, we included 32 testudinid species (all genera and subgenera, and all species of Geochelone, representing 65% of the total familial species diversity), and both mitochondrial (12S rrna, 16S rrna, and cytb) and nuclear (Cmos and Rag2) DNA data with a total of 3387 aligned characters. Using diverse phylogenetic methods (Maximum Parsimony, Maximum Likelihood, and Bayesian Analysis) congruent support is found for a well-resolved phylogeny. The most basal testudinid lineage includes a novel sister relationship between Asian Manouria and North American Gopherus. In addition, this phylogeny supports two other major testudinid clades: Indotestudo + Malacochersus + Testudo; and a diverse clade including Pyxis, Aldabrachelys, Homopus, Chersina, Psammobates, Kinixys, and Geochelone. However, we Wnd Geochelone rampantly polyphyletic, with species distributed in at least four independent clades. Biogeographic analysis based on this phylogeny is consistent with an Asian origin for the family (as supported by the fossil record), but rejects the long-standing hypothesis of South American tortoises originating in North America. By contrast, and of special signiwcance, our results support Africa as the ancestral continental area for all testudinids except Manouria and Gopherus. Based on our systematic Wndings, we also propose modiwcations concerning Testudinidae taxonomy Elsevier Inc. All rights reserved. Keywords: Testudinidae; Tortoises; Systematics; Taxonomy; Biogeography; 12S; 16S; cytb; Cmos; Rag2 1. Introduction The turtle family Testudinidae includes 49 living species of tortoises in 12 extant genera, all of which are completely terrestrial, and which together represent about 18% of the extant world turtle diversity (Ernst and Barbour, 1989; Uetz, 2005). Testudinids are the most broadly distributed non-marine turtle family, occurring on all subpolar continents except Australia, and occupying a wide diversity of habitats varying from rain forests in Southeast Asia and South America, to deserts in North America and Africa. Morphologically, tortoises also * Corresponding author. Fax: address: minhl@amnh.org (M. Le). show a great diversity of sizes and forms. The largest tortoises, in the Galápagos (Geochelone nigra), can reach 1.5 m in length, yet the speckled padloper (Homopus signatus) is mature at only 10cm in length. Most genera have highly domed and rigid carapaces, but hinged-back carapaces occur in the genus Kinixys, and the genus Malacochersus has a remarkable Xattened and Xexible carapace for occupying rock crevices (Crumly, 1984a; Ernst and Barbour, 1989). Despite previous research, phylogenetic relationships within the family Testudinidae have remained controversial (Caccone et al., 1999a; Crumly, 1982, 1984a; Gerlach, 2001, 2004; Meylan and Sterrer, 2000; Parham et al., 2006). One of the perceived problems, voiced by several authors, concerns the high level of morphological convergences suspected for some traditionally used /$ - see front matter 2006 Elsevier Inc. All rights reserved. doi: /j.ympev
2 518 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) characters (AuVenberg, 1974; Bramble, 1971; Pritchard, 1994). In addition, all testudinid molecular studies to date have been limited in terms of taxonomic sampling because they addressed speciwc questions of relationship within smaller subsets of the family (with the possible exception of Cunningham s (2002) study, but this has not yet become available). These studies included relationships within the genus Gopherus (Lamb and Lydeard, 1994), origin and relationships of Malagasy tortoises (Caccone et al., 1999a), origin of Galápagos tortoises (Caccone et al., 1999b), origin of Indian Ocean tortoises (Austin and Arnold, 2001; Austin et al., 2003; Palkovacs et al., 2003), relationships within the genus Indotestudo (Iverson et al., 2001), and relationships within the genus Testudo (Fritz et al., 2005; Parham et al., 2006; van der Kuyl et al., 2002). Nevertheless, the monophyly of the family has been continuously supported by both molecular and morphological studies; with two groups: Geoemydidae and Emydidae considered sister to testudinids (Crumly, 1984a; GaVney and Meylan, 1988; Gerlach, 2001; Krenz et al., 2005; Near et al., 2005; Spinks et al., 2004; van der Kuyl et al., 2002). Within the Testudinidae, establishing the monophyly of the genus Geochelone has proven to be especially problematic. As the largest genus of the family, this group currently includes 10 species, exclusive of Wve species recently assigned to the genera Manouria and Indotestudo based on the studies of Crumly (1984a,b) and Hoogmoed and Crumly (1984), and six extant or recently extinct species being placed in the genus Aldabrachelys (Dipsochelys) (Austin et al., 2003; Bour, 1982; Gerlach, 2001). AuVenberg (1974) proposed six subgenera (including Manouria and Indotestudo) for Geochelone based largely on zoogeographic criteria, however, Crumly (1982) was the Wrst to argue against their complete use based on his cladistic analysis using 26 cranial characters. This view has also been corroborated by more recent analyses, which have included more characters and taxa (Crumly, 1984a; Gerlach, 2001). Crumly s (1984a) results also did not support Geochelone monophyly (Fig. 1). More recent morphological studies have provided conxicting results: GaVney and Meylan (1988) and Meylan and Sterrer (2000) found support for a monophyletic Geochelone, and Gerlach (2001) found Geochelone to be polyphyletic. In addition, recent molecular studies have shown Malagasy Geochelone paraphyletic with respect to Pyxis (Caccone et al., 1999a; Palkovacs et al., 2002), or provided little resolution between Geochelone and other testudinid genera (van der Kuyl et al., 2002). All of these molecular studies were restricted to mitochondrial genes and only included a subset of Geochelone species. In terms of their biogeographic history, tortoises over an interesting group for study because of their almost worldwide distribution, including oceanic islands, e.g., the Galápagos, the East Indies, the West Indies, Madagascar, and the Mascarene Islands. Crumly s (1984a) study has provided the most detailed discussion of diverent biogeographic scenarios of testudinids to date. He suggested true tortoises originated in North America, based on the occurrence of the oldest known fossil testudinid, Hadrianus majusculus, from the Early Eocene. Crumly (1984a) also proposed that tortoises from Europe and Africa were more closely related to Asian forms than to North American forms, and that tortoises of the Galápagos, and the Indian Ocean had rafted to these islands. This latter dispersal scenario has been supported by more recent studies (Austin and Arnold, 2001; Caccone et al., 1999b; Palkovacs et al., 2002). However, the origin of the South American tortoises remains a matter of controversy. Most authors have concluded that tortoises invaded tropical South America from North or Central America (AuVenberg, 1971; Gerlach, 2001; Simpson, 1943; Williams, 1950); however, both Simpson (1942) and Crumly (1984a) also suggested Africa as a possible alternative source. The major objective of our study was to produce a phylogeny of the Testudinidae based on an expanded molecular character dataset, which would be comprehensive at the generic level, and include all the species available to us (the majority of species). We ultimately were able to include species representing all recognized and previously proposed extant genera and subgenera, representing in total 65% of all testudinid species. This broad taxonomic sampling was considered especially important for resolving phylogenetic relationships within the problematic genus Geochelone, and for this group, we were able to include all Geochelone species. To provide a robust estimate of phylogeny, we included a broad diversity of genes, including one mitochondrial protein coding gene (cytb), two mitochondrial ribosomal genes (12S and 16S), and two nuclear genes (Cmos and Rag2), the latter of which have not been previously utilized for resolving relationships between turtles. 2. Materials and methods 2.1. Taxonomic sampling For our testudinid ingroup, we included 34 taxa (32 species and 2 subspecies) and all genera within the family. We also included all Geochelone species. For two species: Indotestudo forstenii and Gopherus agassizii, for which we were unable to obtain tissue; sequences for these species (reported by Spinks et al., 2004) were obtained from Gen- Bank. A complete list of all tissues, voucher specimens, and sequences are provided in Table 1. This molecular phylogenetic analysis includes the most comprehensive taxon sampling achieved to date for testudinids. For outgroups, we included two species of Geoemydidae (Rhinoclemmys melanosterna and R. nasuta) and two species of Emydidae (Glyptemys insculpta and Deirochelys reticularia) based on their reported sister relationships to testudinids recovered by both morphological and molecular evidence (GaVney and Meylan, 1988; Honda et al., 2002; Krenz et al., 2005; Near et al., 2005; Spinks et al., 2004).
3 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) Fig. 1. Crumly s (1984a) preferred hypothesis of relationships for the family Testudinidae, based on 57 morphological characters. In contrast to all the other genera shown here, the genus Geochelone is paraphyletic Molecular data Both mitochondrial and nuclear DNA were utilized in this study to resolve relationships at all phylogenetic levels within the family. Similar approaches have proved successful with other groups exhibiting similar levels of phylogenetic diversity (see Barker et al., 2004; Georges et al., 1999; Hillis et al., 1996; Pereira et al., 2002; Saint et al., 1998). We sequenced three regions of the mitochondrial DNA (mtdna): the complete cytochrome b sequence, and sections of both the 12S and 16S rrna genes. For some taxa (see Table 1), we downloaded cytb and 12S sequences from GenBank originating from studies by Spinks et al. (2004), Palkovacs et al. (2002), Caccone et al. (1999a,b), and van der Kuyl et al. (2002). We also sequenced two nuclear fragments of the Cmos and Rag2 genes. All primers used for this study (including those developed during the course of this study) are shown in Table 2.
4 520 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) Table 1 GenBank accession numbers, and associated voucher specimens/tissues that were used in this study Species names GenBank No. (12S) GenBank No. (16S) GenBank No. (cytb) GenBank No. (Cmos) GenBank No. (Rag2) Voucher numbers for this study Chersina angulata DQ DQ DQ DQ DQ AMCC Aldabrachelys arnoldii AY AY DQ DQ DQ YU Aldabrachelys dussumieri DQ DQ DQ DQ DQ YU Aldabrachelys hololissa AY AY DQ DQ DQ YU Geochelone carbonaria AB AF DQ DQ DQ AMCC Geochelone chilensis AF AF DQ DQ DQ AMCC Geochelone denticulata DQ AF DQ DQ DQ AMCC Geochelone elegans AY AY DQ DQ DQ AMCC Geochelone nigra AF AF DQ DQ DQ FMNH Geochelone p. pardalis DQ DQ DQ DQ DQ AMCC Geochelone p. babcocki DQ DQ DQ DQ DQ AMCC Geochelone platynota DQ DQ DQ DQ DQ AMCC Geochelone radiata AF AF DQ DQ DQ AMCC Geochelone sulcata AY AY DQ DQ DQ AMCC Geochelone yniphora AF AF DQ DQ DQ AMCC Gopherus agassizii a AY AY Gopherus polyphemus AF AF DQ DQ DQ FMNH Homopus boulengeri DQ DQ DQ DQ DQ AMCC Homopus signatus DQ DQ DQ DQ DQ AMCC Indotestudo elongata DQ DQ DQ DQ DQ AMCC Indotestudo forstenii a AY Indotestudo travancorica DQ DQ DQ DQ DQ AMCC Kinixys belliana DQ DQ DQ DQ DQ AMCC Kinixys homeana DQ DQ DQ DQ DQ KU Malacochersus tornieri DQ DQ DQ DQ DQ AMCC Manouria emys emys DQ DQ DQ DQ DQ AMCC Manouria emys phayrei DQ DQ DQ DQ DQ AMCC Manouria impressa DQ DQ DQ DQ DQ AMCC Psammobates tentorius DQ DQ DQ DQ DQ AMCC Pyxis arachnoides AF AF DQ DQ DQ RAN34680 Pyxis planicauda AF AF DQ DQ DQ AMCC Testudo graeca AF DQ DQ DQ DQ AMCC Testudo horsweldii AB DQ DQ DQ DQ AMCC Testudo kleinmanni AF DQ DQ DQ DQ AMCC Glyptemys insculpta DQ DQ AF DQ DQ AMCC Deirochelys reticularia DQ DQ AF DQ DQ AMCC Rhinoclemmys melanosterna DQ DQ AY DQ DQ AMCC Rhinoclemmys nasuta DQ DQ DQ DQ DQ AMCC All sequences generated by this study have accession numbers: DQ DQ497396; YU, Yale University Collection; FMNH, Field Museum of Natural History; AMCC, Ambrose Monell Cryo Collection, American Museum of Natural History ( RAN, Ronald A. Nussbaum Field Series, University of Michigan, Museum of Zoology; KU, Kansas University Natural History Museum. a Genbank sequences only. DNA was extracted from tissues and blood samples using the DNeasy kit (Qiagen) following manufacturer s instructions for animal tissues. PCR volume for mitochondrial genes contained 42.2 μl (18 μl water, 4 μl of buver, 4 μl of 20 mm dntp, 4 μl of 25 mm MgCl 2, 4 μl of each primers, 0.2 μl of Taq polymerase (Promega), and 4 μl of DNA). PCR conditions for these genes were: 95 C for 5 min to activate the Taq; with 42 cycles at 95 C for 30 s, 45 C for 45 s, 72 C for 60 s; and a Wnal extension of 6 min. Nuclear DNA was ampliwed by HotStar Taq Mastermix (Qiagen) since this Taq performs well on samples with low-copy targets and the Taq is highly speciwc. The PCR volume consists of 21 μl (5 μl water, 2 μl of each primer, 10 μl of Taq mastermix, and 2 μl of DNA or higher depending on the quantity of DNA in the Wnal extraction solution). PCR conditions for nuclear genes were the same as above except the Wrst step (95 C) was carried out in 15 min, and the annealing temperatures were 52 and 58 C for Rag2 and Cmos, respectively. PCR products were visualized using electrophoresis through a 2% low melting-point agarose gel (NuSieve GTG, FMC) stained with ethidium bromide. For reampliwcation reactions, PCR products were excised from the gel using a Pasteur pipette, and the gel plug was melted in 300 μl sterile water at 73 C for 10 min. The resulting gel-puriwed product was used as a template in 42.2μl reampliwcation reactions with all PCR conditions similar to those used for mitochondrial genes. PCR products were cleaned using PerfectPrep PCR Cleanup 96 plate (Eppendorf) and cycle sequenced using ABI prism big-dye terminator according to manufac-
5 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) Table 2 Primers used in this study Primer Position Sequence Reference L1091 (12S) AAAAAGCTTCAAACTGGGATTAGATACCCCACTAT-3 Kocher et al. (1989) H1478 (12S) TGACTGCAGAGGGTGACGGGCGGTGTGT-3 Kocher et al. (1989) AR (16S) CGCCTGTTTATCAAAAACAT-3 Palumbi et al. (1991) BR (16S) CCGGTCTGAACTCAGATCACGT-3 Palumbi et al. (1991) CytbG (cytb) AACCATCGTTGTWATCAACTAC-3 Spinks et al. (2004) GLUDGE (cytb) TGATCTTGAARAACCAYCGTTG-3 Palumbi et al. (1991) CytbJSi (cytb) GGATCAAACAACCCAACAGG-3 Spinks et al. (2004) CytbJSr CCTGTTGGGTTGTTTGATCC-3 Spinks et al. (2004) THR (cytb) TCATCTTCGGTTTACAAGAC-3 Spinks et al. (2004) THR-8 (cytb) GGTTTACAAGACCAATGCTT-3 Spinks et al. (2004) CM1 (Cmos) GCCTGGTGCTCCATCGACTGGGA-3 Barker et al. (2002) CM2 (Cmos) GGGTGATGGCAAAGGAGTAGATGTC-3 Barker et al. (2002) Cmos1 (Cmos) GCCTGGTGCTCCATCGACTGGGATCA-3 This study Cmos3 (Cmos) GTAGATGTCTGCTTTGGGGGTGA-3 This study F2 (Rag2) CAGGATGGACTTTCTTTCCATGT-3 a This study F2-1 (Rag2) TTCCAGAGCTTCAGGATGG-3 This study R2-1 (Rag2) CAGTTGAATAGAAAGGAACCCAAGT-3 b This study a ModiWed from F2R2 (Barker et al., 2004). b ModiWed from R2R1 (Barker et al., 2004); Cmos and Rag2 primer positions corresponding to the positions in chicken genomes with GenBank numbers of M19412 and M58531, respectively; primer positions for mitochondrial genes corresponding to the positions in the complete mitochondrial genome of Chrysemys picta (Mindell et al., 1999). turer recommendation. Sequences were generated in both directions on an ABI 3100 Genetic Analyzer Phylogenetic analysis We aligned sequence data using ClustalX v1.83 (Thompson et al., 1997) with default settings. Data were analyzed using both parametric and non-parametric methods (see Kolaczkowski and Thornton, 2004): maximum parsimony (MP) and maximum likelihood (ML) using PAUP*4.0b10 (SwoVord, 2001) and Bayesian analysis using MrBayes v3.1 (Huelsenbeck and Ronquist, 2001). For maximum parsimony analysis, we conducted heuristic analyses with 100 random taxon addition replicates using the tree-bisection and reconnection (TBR) branch swapping algorithm in PAUP, with no upper limit set for the maximum number of trees saved. Bootstrap support (BP) (Felsenstein, 1985a) was evaluated using 1000 pseudoreplicates and 100 random taxon addition replicates. Bremer indices (BI) (Bremer, 1994) were determined using Tree Rot 2c (Soreson, 1999). All characters were equally weighted and unordered. Gaps in sequence alignments were treated as a Wfth character state (Giribert and Wheeler, 1999). Phylogenetic congruence between partitions of the nuclear and mitochondrial datasets was assessed using the incongruence length diverence (ILD) test (Farris et al., 1994). Although the utility of this test as a measure for data combination has been questioned (e.g., see Darlu and Lecointre, 2002; Yoder et al., 2001), we employed this test to further explore characteristics of our partitioned data, along with phylogenetic analyses that we also conducted using these same partitions. These data were partitioned by genes and DNA organelle (nuclear or mitochondrial), and also analyzed in their entirety. For maximum likelihood analysis the optimal model for nucleotide evolution was determined using Modeltest V3.7 (Posada and Crandall, 1998). Analyses used a randomly selected starting tree, and heuristic searches with simple taxon addition and the TBR branch swapping algorithm. Support for the likelihood hypothesis was evaluated by bootstrap analysis with 100 replications and simple taxon addition. For Bayesian analyses we used the optimal model determined using Modeltest with parameters estimated by MrBayes Version 3.1. Analyses were conducted with a random starting tree and run for generations. Four Markov chains, one cold and three heated (utilizing default heating values), were sampled every 1000 generations. Log-likelihood scores of sample points were plotted against generation time to detect stationarity of the Markov chains. Trees generated prior to stationarity were removed from the Wnal analyses using the burn-in function. Two independent analyses were started simultaneously. The posterior probability values (PP) for all clades in the Wnal majority rule consensus tree are reported. We ran analyses on both combined and partitioned datasets to examine the robustness of the tree topology (Brandley et al., 2005; Nylander et al., 2004). In the partitioned analyses, we divided the data into 11 separate partitions, including 12S, 16S, and the other nine based on gene codon positions (Wrst, second, and third) in cytb, Cmos, and Rag2. Optimal models of molecular evolution for each partition were selected using Modeltest and then assigned to these partitions in MrBayes 3.1 using the command APPLYTO. Model parameters were estimated independently for each data partition using the UNLINK command. To test alternative hypotheses of relationships, corresponding tree topologies were compared using the Wilcoxon signed-ranks test (Felsenstein, 1985b; Templeton, 1983), to determine if tree length diverence could have resulted from chance alone (Larson, 1998). Alternative tree topologies were
6 522 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) Fig. 2. The maximum parsimony tree based on all data combined. This is the strict consensus of two most parsimonious trees, (TL D 4292; CI D 0.39; RI D 0.50). The combined Wve-gene dataset includes 3387 aligned characters of which 880 are potentially parsimony informative. Numbers above and below branches are bootstrap (>50%) and Bremer values, respectively. The geographic distribution of each taxon is indicated in the right column. constructed in MacClade (Maddison and Maddison, 2001) and then used as constraint trees by importing to PAUP. To determine biogeographic support for ancestral areas or vicariance events between clades, we utilized Dispersal-Vicariance Analysis (Ronquist, 1997), as implemented in the program DIVA 1.1 (Ronquist, 1996). Fully resolved trees based on MP and ML topologies were imported using distribution areas (as depicted in Figs. 2 and 4). All optimization settings used the default, including the maximum number of ancestral areas that were set as the total number of unit areas. 3. Results Sequences for all Wve genes were obtained for each of the 32 taxa with tissues. The Wnal matrix consists of 3387 aligned characters: 12S (408 characters), 16S (583 characters), cytb (1140 characters), Cmos (602 characters), and Rag2 (654 characters). Five taxa have sequences that include gaps at three loci (cytb, Cmos, and Rag2). For cytb, Geochelone pardalis babcockii has a single indel that is three base pairs long at positions For Cmos, G. platynota and G. elegans has a single indel that is six base pairs long corresponding to positions of the complete chicken Cmos sequence (GenBank Accession No. M19412). For Rag2, Chersina angulata and Kinixys homeana has a single indel of three base pairs long corresponding to positions of the partial Pelodiscus sinensis Rag2 sequence (GenBank Accession No. AF369089). The IDL tests found no signiwcant incongruence between mitochondrial genes (12S vs 16S, p D 0.07; cytb vs 12S, p D 0.89; cytb vs 16S, p D 0.97), between nuclear genes (Cmos vs Rag2, p D 0.16), and between the nuclear and mitochondrial partitions (nuclear DNA vs mtdna, p D 0.7). Using MP, we also analyzed the data by gene and organelle partitions (see Tables 3 and 4). The analysis indicated that Clades 3, 4, and 5 (see Fig. 4) consistently receive strong support from both mitochondrial and nuclear genes (Table 3). For the following partitions, the strict consensus trees include little phylogenetic resolution: 12S, 16S, Cmos, Rag2, and all the nuclear data. For cytb, although MP recovered only two equally parsimonious trees, the strict consensus is poorly resolved concerning deeper level relationships between clades including multiple genera. The strict consensus tree using all mtdna was unresolved for
7 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) Table 3 MP analyses of data partitions with BP support values for each clade as dewned in Fig. 4 Data Clade 1 (%) Clade 2 (%) Clade 3 (%) Clade 4 (%) Clade 5 (%) Clade 6 (%) Clade 7 (%) Clade 8 (%) 12S < <50 <50 <50 <50 <50 16S < <50 99 <50 <50 <50 Cytb 55 <50 75 <50 <50 <50 <50 <50 All MtDNA < Cmos <50 < <50 <50 <50 Rag2 <50 <50 <50 92 <50 <50 <50 <50 All Nuclear < <50 <50 <50 Table 4 Data partitions subject to phylogenetic analyses with maximum parsimony Data Total number of aligned sites Parsimony informative characters Variable characters MP tree length 12S S cytb Cmos Rag All MtDNA All Nuclear All data RI value CI value Number of equally parsimonious trees basal nodes of the clade including all the Geochelone groups the same clade as recovered in the combined analysis (see Fig. 2). All nodes with BI D 1 in the combined MP analysis collapsed in this mtdna tree. Based on the generally poorly resolved tree topologies recovered using the partitioned data, and our IDL partition results, we thus consider the combined data as providing the optimal set upon which to base phylogenetic inference. Using the combined dataset (3387 aligned characters, 880 potentially parsimony informative), the MP analysis recovered two most parsimonious trees with 4292 steps (CI D 0.39; RI D 0.50). The strict consensus is shown in Fig. 2. This tree is well resolved with 72% of the nodes receiving strong support (BP 7 70%) (Hillis and Bull, 1993), and 9% receiving BP support from 65 to 69%. All nodes in the clades, including Manouria, Gopherus, Testudo, Indotestudo, and Malacochersus, are well supported, except for the relationship of Testudo horsweldii and Malacochersus. We also found that the BP value supporting the sister relationship between Gopherus and Manouria increases signiwcantly (from 73 to 85% in the MP analysis) by excluding the species with incomplete data, Gopherus agassizii. Within the diverse clade, which includes Geochelone, Aldabrachelys, Kinixys, Homopus, Psammobates, and Pyxis, support was generally high except for some of the basal relationships. Incongruence between these two MP trees was conwned to the genus Aldabrachelys. Results, including model parameters, of the ML and Bayesian analyses are shown in Fig. 3 (tree topologies of both analyses are identical). Similar to the MP analysis, the ML tree is well resolved with 78% of the nodes receiving strong support (BP 7 70%). The most weakly supported nodes are distributed in the clade containing Geochelone. The topology of the ML cladogram is congruent with the MP topology for all branches except the following: Malacochersus tornieri sister to either Testudo (BP D 68%, MP) or Indotestudo (BP D 60%, ML), G. radiata sister to either Pyxis (BP D 65%, MP) or Geochelone yniphora (BP D 82%, ML), and the clade including Kinixys + South American Geochelone (G. carbonaria group) sister to either the clade including the Malagasy and Indian Ocean species, Chersina, Homopus, Psammobates, and G. pardalis (<50%, MP) or the clade including G. elegans, G. platynota, and G. sulcata (87%, ML). In the combined Bayesian analysis (Fig. 3), ln L scores reached equilibrium after 19,000 generations while the partitioned Bayesian analysis scores reached stationarity after 27,000 generations in both runs. Phylogenetic relationships supported by the combined and partitioned data are identical to the ML results. Only minor diverences in the PP values were found between combined and partitioned Bayesian cladograms: PP values are higher in the partitioned analysis for the following clades: Pyxis and Aldabrachelys (99%), South American testudinids (99%), Kinixys + South American testudinids (86%), Testudo (99%), and A. arnoldi + A. hololissa (74%), and lower for the following: Chersina + Homopus + Psammobates + Geochelone pardalis + Aldabrachelys + Pyxis + Geochelone radiata + G. yniphora (56%), G. chilensis + G. nigra (93%), and Malacochersus + Indotestudo (95%). All of our analyses, thus, found three well supported clades of tortoises: (1) Gopherus + Manouria, (2)
8 524 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) Fig. 3. The maximum likelihood (ML) analysis phylogram based on all the data combined, using the GTR+G+I model of molecular evolution (Bayesian tree topology is identical). Base frequency A D 0.313, C D 0.262, G D 0.193, T D ML ln L D ; rate matrix: A C D 3.544, A G D , A T D 3.478, C G D 0.897, C T D , G T D 1.000; proportion of invariable site (I) D 0.470; gamma distribution shape parameter (G) D Total number of rearrangements tried D 13077, score of best tree found D Numbers above and below branches are ML bootstrap values (>50%) and Bayesian posterior probabilities, respectively. See Fig. 2 for taxon distribution. The sister relationship between A. arnoldi and A. hololissa, cannot be seen due to the short branch length. Indotestudo + Testudo + Malacochersus, and (3) Geochelone +Pyxis+Aldabrachelys+Homopus+ Chersina + Psammobates +Kinixys. It is also evident from these analyses that Geochelone, as currently dewned, is rampantly polyphyletic. Our phylogenetic results place Geochelone in at least four independent clades. The Wrst consists of three species, G. elegans, G. platynota, and G. sulcata (G. elegans group). The second clade includes all four South American species (G. carbonaria group), which are sister to Kinixys. G. yniphora and G. radiata from Madagascar belong to a third clade that also includes Pyxis and Aldabrachelys. The Wnal clade includes only a single Geochelone species, G. pardalis, which is sister to the genus Psammobates. Biogeographic results are summarized by the area cladogram shown in Fig. 4, which conservatively depicts the strict congruence between our MP, ML, and Bayesian tree topologies. The optimized ancestral areas, based on DIVA, are also shown in Fig. 4. The DIVA ancestral area for the family Testudinidae is identiwed as including three areas: Asia, Africa, and North America, and requires a further seven subsequent dispersals (to South America, Madagascar Indian Ocean, and Asia). For this three-continent familial ancestral area, Gopherus represents the North American lineage, Manouria represents the Asian lineage, and all other testudinids represent the African lineage. For the incongruent clades between the MP and ML analyses (see Figs. 2 and 3), DIVA ancestral areas are optimized as
9 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) Fig. 4. Testudinidae area cladogram based on the strict consensus of the tree topologies shown in Figs. 2 and 3. The ancestral area optimizations found by dispersal-vicariance analysis (DIVA) are shown for each node: Asia (As), Africa (Af), Madagascar and Indian Ocean (MI), North America (NA), and South America (SA). follows: Malacochersus + Testudo, Africa; Malacochersus + Indotestudo, Africa and Asia; Chersina + Homopus+ Psammobates + Geochelonepardalis + Kinixys +the Geochelone carbonaria group + Aldabrachelys + Pyxis + Geochelone radiata + G. yniphora, Africa; and the G. elegans group + Kinixys +the Geochelone carbonaria group, Africa. 4. Discussion 4.1. Phylogeny Concerning the three major clades of testudinids we Wnd in this study, the sister group to all other tortoise species includes Manouria and Gopherus as sister genera (Figs. 2 and 3). This is the Wrst study to sample broadly across the family and recover this intriguing sister relationship; although Lamb and Lydeard s (1994) molecular study also found weak support for this sister relationship. Other morphological and molecular studies have consistently reported Manouria to be basal to all other tortoises (Crumly, 1984a; GaVney and Meylan, 1988; Gerlach, 2001; Meylan and Sterrer, 2000; Takahashi et al., 2003; Spinks et al., 2004), with Gopherus usually placed as the next most basal lineage for the family (Crumly, 1984a; Spinks et al., 2004). Manouria and Gopherus are distributed in Asia and North America, respectively, and these two genera show obvious diverences in morphological characters, with Manouria exhibiting several character states considered to be plesiomorphic, such as the wide anterior edge of the prootic bone (ranging from 80 to 92% of the posterior edge) and the ventrally and dorsally split supracaudal scute (Crumly, 1982). This Manouria + Gopherus sister relationship has, to the best of our knowledge, not been previously considered or discussed, and no synapomorphies have been proposed for the two genera (GaVney and Meylan, 1988; Gerlach, 2001; Meylan and Sterrer, 2000; Takahashi et al., 2003). Winokur and Legler (1975) reported mental glands in Gopherus, and Ernst and Barbour (1989), also citing Winokur and Legler, report mental glands in Manouria thus suggesting a potentially homologous character. However, our reading of Winokur and Legler s (1975) study clearly reveals that for testudinids, these authors only reported mental glands for Gopherus. In addition, our own examination of AMNH catalogued material (R and R ), and living specimens, also failed to Wnd mental glands in Manouria. The second major clade found by this study includes Testudo, Indotestudo, and Malacochersus. A recent molecular study by Parham et al. (2006) based on the complete mitochondrial genome also recovered this monophyletic group. Their MP, ML, and Bayesian analyses produced conxicting results regarding the relationships of Malacochersus tornieri and Testudo horsweldii. SpeciWcally, their MP and Bayesian analyses supported the sister relationship
10 526 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) between Malacochersus and Indotestudo, but the ML analysis found Malacochersus sister to all other taxa. The position of T. horsweldii was unresolved in the MP analysis (strict consensus tree), but was sister to T. hermanni in the other analyses. Another study (based on 1124 bp of cytb) by Fritz et al. (2005) which included almost all forms of the genus Testudo plus Indotestudo and Malacochersus was also ambiguous regarding relationships among these three genera. Similarly, we also found conxicting branch support for relationships within this clade: the MP analysis give moderate support (BP D 68%) for a Malacochersus + Testudo clade, while the ML and Bayesian results supported a Malacochersus + Indotestudo clade (ML BP D 60%, PP D 97%). Interestingly, morphological studies have consistently supported Malacochersus and Testudo as sisters (Crumly, 1984a; GaVney and Meylan, 1988; Gerlach, 2001). According to Gerlach (2001), the two genera share the character of the processus inferior parietalis meeting the quadrate and partially covering the prootic, while GaVney and Meylan (1988) found they share four characters: presence of supranasal scales, no contact between the inguinal and femoral scutes, a viewable ventral tip of the processus interfenestralis, and the presence of sutures between this process and the surrounding bones. Our dataset unambiguously resolves relationships within the three Indotestudo species. The sister relationship between I. travancorica and I. elongata is recovered by MP, ML, and Bayesian analyses with strong support. In previous studies, relationships among these species had remained uncertain. The morphological study by Hoogmoed and Crumly (1984) found I. travancorica and I. forstenii to be identical, yet the molecular study by Iverson et al. (2001) found strong support for the sister relationship between I. travancorica and I. elongata using MP, but their ML analysis favored I. travancorica and I. forstenii as sister taxa. The study by Spinks et al. (2004) also supported this latter hypothesis. For the third and largest testudinid clade, we Wnd support for Wve internal clades: (1) Chersina + Homopus + Psammobates+ Geochelone pardalis, (2) the Geochelone elegans group, (3) Kinixys, (4) the Geochelone carbonaria group, and (5) Aldabrachelys + Pyxis+ Geochelone radiata and G. yniphora. However, relationships between these clades con- Xict between the MP, ML, and Bayesian analyses, and corresponding branch support is also generally lower (MP BP<50%, ML BP<50 87%, and Bayesian PP %). Of special signiwcance (and as suspected by previous researchers), we Wnd substantial polyphyly for the problematic genus Geochelone. Nevertheless, the phylogenetic results we present here for Geochelone were not recovered by previous morphological and molecular studies (Gerlach, 2001; Meylan and Sterrer, 2000; van der Kuyl et al., 2002; Palkovacs et al., 2002). The strongly supported sister relationships we Wnd between G. pardalis and Psammobates, between Chersina and Homopus; and the monophyly of Geochelone elegans group (G. elegans, G. platynota, and G. sulcata) were not evident in previous morphological studies; and the molecular study by Palkovacs et al. (2002) found support for conxicting relationships concerning G. sulcata and G. elegans between MP, ML, Bayesian, and neighbor-joining analyses. Although only weakly supported in terms of branch support (MP and ML BP < 50%, PP 80%), all of our analyses support the intriguing sister relationship between African Kinixys and the South American Geochelone carbonaria group. This hypothesis of relationship is radical: prior morphological and molecular studies placed members of the Geochelone carbonaria group either sister to Gopherus (Crumly, 1984a; Gerlach, 2001), or sister to other Geochelone species: G. elegans, G. pardalis, and G. sulcata (Crumly, 1984a; Meylan and Sterrer, 2000; Palkovacs et al., 2002; van der Kuyl et al., 2002). Crumly (1984a) did not consider the South American tortoises to be monophyletic (Fig. 1), but other researchers have supported monophyly for a sampled subset of this clade (van der Kuyl et al., 2002), or else otherwise explicitly assumed it (Caccone et al., 1999b; GaVney and Meylan, 1988; Gerlach, 2001; Meylan and Sterrer, 2000). The Indian Ocean clade supported by our results: Aldabrachelys + Pyxis + Geochelone radiata + G. yniphora is consistent with the molecular results of Austin and Arnold (2001) and Palkovacs et al. (2002). The latter study, like ours, found support for a sister relationship between G. radiata and Pyxis using MP (65% BP), and an alternative sister relationship of G. radiata and G. yniphora using Bayesian and ML analyses (ML 82% BP, 100% PP). Morphologically, the sister relationship between G. yniphora and G. radiata has been frequently supported (e.g., Crumly, 1984a; GaVney and Meylan, 1988; Gerlach, 2001; Meylan and Sterrer, 2000) and Gerlach (2001) reported two synapomorphies: a ventral ridge on the maxilla premaxilla suture and keels on the supraoccipital crest. In addition, the close relationship between Pyxis and G. yniphora + G. radiata was also supported by Gerlach with the synapomorphy of an indistinct fenestra postotica. By contrast, other studies (e.g., GaVney and Meylan, 1988; Meylan and Sterrer, 2000; Takahashi et al., 2003) have varied widely concerning the relationship of Pyxis to other testudinid genera. Concerning the Indian Ocean giant tortoises (Aldabrachelys), for all Wve genes we found almost no variation among the three forms: A. dussumieri, A. arnoldi, and A. hololissa. Only two sites showed variation: one for 16S (position 2004) and the other for Cmos (position 349). These results support the conclusions of Palkovacs et al. (2003) and Austin et al. (2003) that all three forms may actually represent a single species and population originating from Aldabra. However, these molecular results need to be evaluated by carefully examining the diagnostic morphological characters used to describe these species in order to conwrm that these features do not vary intraspeciwcally IntraspeciWc divergence The two samples representing diverent subspecies of Geochelone pardalis: G. p. pardalis and G. p. babcockii have
11 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) an uncorrected pairwise sequence divergence distance of 5.9% for cytb, the fastest evolving gene in this study with the highest proportion of variable sites (Table 4). These two subspecies are also known to exhibit morphological diverences. The carapace shape of G. p. babcockii is distinctly convex dorsally, compared to the Xatter G. p. pardalis carapace; and juvenile G. p. babcockii has single carapacial scute spots, while G. p. pardalis has two (Loveridge and Williams, 1957). Furthermore, there is a sexually dimorphic size diverence: G. p. pardalis males being slightly larger than females and G. p. babcockii males being much smaller (Broadley, 1989). G. p. pardalis occurs in western South Africa and southern Namibia. G. p. babcockii ranges from Sudan and Ethiopia to Natal and from Cape Province to southwest Africa and southern Angola (Iverson, 1992; Loveridge and Williams, 1957). Our preliminary results suggest that a phylogeographic study of G. pardalis is expected to reveal high genetic divergence within this species, or species complex. By contrast, the cytb uncorrected pairwise sequence divergence distance between the two samples we have of Manouria e. emys (Burmese tortoise) and M. e. phayrei (Burmese black tortoise) is just 1.3%. Results showing even less geographic variation for mtdna within a testudinid species have been recently reported for Geochelone radiata (Leuteritz et al., 2005) Biogeographic history The earliest Testudinidae fossils (not yet described) are reported to be from the lower Paleocene of Asia (Claude and Tong, 2004; Hutchison, 1998; Joyce et al., 2004), however older fossils of the putative ancestor (family Lindholmemydidae) for the superfamily Testudinoidea are known from the upper Cretaceous of Asia (Nessov, 1988; Sukhanov, 2000). The appearance of testudinids fossils by the early Eocene in both Europe (Achilemys cassouleti) and North America (Manouria or Manouria-like fossils and Hadrianus majusculus), and by the late Eocene in Africa (Gigantochersina ammon) (Claude and Tong, 2004; Crumly, 1984a; Holroyd and Parham, 2003; Hutchison, 1996, 1998), indicates that if tortoises Wrst originated in Asia, they must have soon dispersed to other continents. The abundance of fossil testudinids also suggests that the fossil record is unlikely to be substantially underestimating the origin time for the family. All evidence to date, thus, supports a late Mesozoic or early Cenozoic origin for the Testudinidae in Asia, with dispersal to North America, Europe, and Africa having been achieved by the Eocene. Our DIVA results also establish the ancestral area for testudinids as including three continents: Asia, North America, and Africa, and Wnds Manouria and Gopherus as the extant groups representing the Asian and North American lineages, respectively. Remarkably, all other extant testudinid groups are associated with the African ancestral area. The remarkable biographic basal lineage bifurcation between clades distributed in Asia and North America, which we Wnd here for Manouria and Gopherus, has also been reported in the family Dibamidae (blind skinks) and the skink genus Scincella, which are distributed in the Americas and Asia (Honda et al., 2003; Zug et al., 2001). These Asian American divergences may have resulted from the well-known disruption of Xoral and faunal exchange across the Bering Strait in the early Eocene; a biogeographic pattern that has been well documented in other groups (Beard, 2002; Bowen et al., 2002; Maekawa et al., 2005; Sanmartin et al., 2001 and references therein; TiVney, 1985). The DIVA ancestral area optimization results provide a striking conclusion: it is most parsimonious to consider all testudinids with the exception of Manouria and Gopherus as having an African ancestral area. The alternative biogeographic scenarios, such as considering the ancestral area for this large group as Asia or the Palearctic, will require making additional assumptions of dispersal or extinction. Our results infer the following dispersals from Africa: Indotestudo and the clade including Geochelone platynota + G. elegans dispersing into Asia, Testudo dispersing into Europe, the clade including Aldabrachelys + Pyxis + Geochelone radiata + G. yniphora dispersing into the Indian Ocean, and the G. carbonaria group dispersing into South America. Africa should therefore be viewed as both a major center of origin and diversiwcation for testudinid species. Three clades found by our study are endemic to Africa: Malacochersus, Kinixys, and the diverse clade including Homopus + Chersina + Psammobates + Geochelone pardalis, and these groups presumably diversiwed on this continent (the Kinixys belliana population of Madagascar is typically considered as introduced, see Raselimnanana and Vences, 2003). The dispersal of Testudo and Indotestudo from Africa into Europe and Asia appears to be a novel biogeographic scenario that has not been previously considered. For example, Parham et al. (2006) only discuss dispersal events from a Palearctic center of diversiwcation for Testudo, Indotestudo, and Malacochersus. In addition, Crumly s (1984a) discussion of biogeographic scenarios implied that African tortoises originated from Asia. However, and by contrast, the Indian Ocean clade including Aldabrachelys + Pyxis + Geochelone radiata + G. yniphora has been previously proposed to have an African origin (Caccone et al., 1999a; Palkovacs et al., 2002), as supported by our DIVA ancestral area optimizations (Fig. 4). However, perhaps the most unusual biogeographic result concerns the South American + Galápagos tortoises (Geochelone carbonaria group), which we Wnd are members of a larger clade otherwise only including species from Africa, the Indian Ocean, and Asia. To test the previously proposed North American origin for the Geochelone carbonaria clade, based upon a sister relationship with Gopherus (e.g., Gerlach, 2001) we searched for the shortest constraint tree that included these clades as sisters. This constraint tree was 48 steps longer than our most parsimonious hypothesis of relationship, and this step length
12 528 M. Le et al. / Molecular Phylogenetics and Evolution 40 (2006) diverence was signiwcant (p < 0.001). By contrast, the consistently recovered sister relationship we found between African Kinixys and the Geochelone carbonaria clade, and the distribution of the group sister to this clade, which also includes Africa, providing strongest support for an African origin for the South American tortoises (Fig. 4). The DIVA ancestral area optimizations Wnd this entire clade (Geochelone + Pyxis + Aldabrachelys + Homopus + Chersina + Psammobates + Kinixys) to have an African ancestral area, thus supporting an early evolutionary history for this diverse tortoise group in this continent. The dispersal of tortoises from Africa to South America might have been aided by westward sea currents, such as the Equatorial Currents (Fratantoni et al., 2000). These Xow between the Congo delta and Maranhao in Brazil and are believed to have been in this direction since the breakup of Gondwana (Houle, 1999; Parrish, 1993). Several vertebrate animal groups are reported to show a pattern of dispersal from Africa to South America across the Atlantic: platyrrhine monkeys and caviomorph rodents between 85 and 35 MYA, (Houle, 1999; Huchon and Douzery, 2001; Mouchaty et al., 2001; Nei et al., 2001; Schrago and Russo, 2003), and Mabuya skinks twice during the last 9 million years (Carranza and Arnold, 2003). Tortoises are well known for their ability to Xoat (while extending their heads above water) and survive without food or freshwater for up to six months (Caccone et al., 2002), and oceanic dispersal by testudinid tortoises appears to have occurred many times. For example: Geochelone to the West Indies (Williams, 1950, 1952), Geochelone to the Galápagos and at least Wve subsequent dispersals between islands in the archipelago (Caccone et al., 1999b, 2002; Russello et al., 2005), ancestral Malagasy tortoises to Madagascar from mainland Africa (Caccone et al., 1999a), Aldabrachelys to Aldabra, Astove, Cosmoledo, Denis, Amirantes, Comoros, Assumption, and the Granitic Seychelles (Austin et al., 2003); Cylindraspis (now extinct) to Réunion, Mauritius, and Rodrigues (Austin and Arnold, 2001); and Hesperotestudo (now extinct) to Bermuda (Meylan and Sterrer, 2000) Taxonomic issues Based on our phylogenetic results, and to rexect monophyletic groupings within the Testudinidae, we propose the following taxonomic changes: (1) The name Geochelone (Fitzinger, 1835; type species, G. elegans) is retained for just three species: G. sulcata, G. elegans, and G. platynota. (2) The South American and Galápagos Island species (Geochelone carbonaria group) are placed in the genus Chelonoidis (Fitzinger, 1835; type species, C. carbonaria), which has been previously recognized as a subgenus (Fitzinger, 1856). (3) Geochelone pardalis is placed in the genus Psammobates (Fitzinger, 1835). (4) Based on the desirability of maintaining the morphologically distinct and well-supported genera Pyxis and Aldabrachelys, yet also due to the uncertainty concerning the relationships of Geochelone yniphora and G. radiata to these genera, we therefore prefer to conservatively place both these species in separate monotypic genera. We recommend the use of the available genus name Astrochelys (Gray, 1873; type species, A. radiata) for G. radiata. However, because there is no available generic name for G. yniphora, we thus propose the following new genus: Angonoka new genus Type species. Testudo yniphora Vaillant Diagnosis Testudinid tortoises that have the following characters: (1) gulars fused to form a single scute; (2) gular projection thickened and sexually dimorphic in size, projecting well beyond the carapace edge and curved upward in adult males; (3) highly domed carapace with descending sides; (4) carapace brown, lacking radiating yellow or black lines on scutes; (5) no enlarged tubercles on thighs; (6) tail without an enlarged terminal scale; (7) longitudinal ventral ridge on the maxilla premaxilla suture, (8) horizontal keels on the supraoccipital crest; (9) pectoral scutes narrow and not contacting the entoplastron. Content. One species, Angonoka yniphora (Vaillant 1885). Distribution. Northwest Madagascar. Etymology. The generic name Angonoka is based on the Malagasy word Angonoka which is the local name for the type species. 5. Conclusions The broad taxonomic sampling of tortoises we were able to achieve with this study, coupled with the inclusion of both mitochondrial and nuclear genes, has resulted in supported tree topologies with both good resolution, and for many branches, high levels of support. Importantly, these topologies are also largely congruent, regardless of the approach taken concerning phylogenetic analysis. We consider this result to represent a substantial advance in our understanding of testudinid relationships, especially concerning the problematic genus Geochelone, which has been in a state of substantial taxonomic confusion for the past 30 years. Although our results Wnd Geochelone polyphyletic, representing at least four independent clades, we anticipate these Wndings will stimulate new interest within these tortoise groups and strengthen future comparative research. Despite these systematic advances, further phylogenetic analysis will be needed to test the relationships we report here between Indotestudo, Testudo, and Malacochersus, and the relationships of G. yniphora and G. radiata. The inclusion of additional genes (nuclear and mitochondrial) and morphological data will likely resolve this uncertainty. Importantly, a re-examination of the morphological characters will also provide new insights concerning character evolution for the group, and in addition, aid with the interpretation of the fossil taxa. Unlike most extant reptile families, tortoises have a comparably rich Cenozoic fossil
On the paraphyly of the genus Kachuga (Testudines: Geoemydidae)
 Molecular Phylogenetics and Evolution 45 (2007) 398 404 Short communication On the paraphyly of the genus Kachuga (Testudines: Geoemydidae) Minh Le a,b, *, William P. McCord c, John B. Iverson d a Department
Molecular Phylogenetics and Evolution 45 (2007) 398 404 Short communication On the paraphyly of the genus Kachuga (Testudines: Geoemydidae) Minh Le a,b, *, William P. McCord c, John B. Iverson d a Department
Lecture 11 Wednesday, September 19, 2012
 Lecture 11 Wednesday, September 19, 2012 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean
Lecture 11 Wednesday, September 19, 2012 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean
Species: Panthera pardus Genus: Panthera Family: Felidae Order: Carnivora Class: Mammalia Phylum: Chordata
 CHAPTER 6: PHYLOGENY AND THE TREE OF LIFE AP Biology 3 PHYLOGENY AND SYSTEMATICS Phylogeny - evolutionary history of a species or group of related species Systematics - analytical approach to understanding
CHAPTER 6: PHYLOGENY AND THE TREE OF LIFE AP Biology 3 PHYLOGENY AND SYSTEMATICS Phylogeny - evolutionary history of a species or group of related species Systematics - analytical approach to understanding
Turtles (Testudines) Abstract
 Turtles (Testudines) H. Bradley Shaffer Department of Evolution and Ecology, University of California, Davis, CA 95616, USA (hbshaffer@ucdavis.edu) Abstract Living turtles and tortoises consist of two
Turtles (Testudines) H. Bradley Shaffer Department of Evolution and Ecology, University of California, Davis, CA 95616, USA (hbshaffer@ucdavis.edu) Abstract Living turtles and tortoises consist of two
CLADISTICS Student Packet SUMMARY Phylogeny Phylogenetic trees/cladograms
 CLADISTICS Student Packet SUMMARY PHYLOGENETIC TREES AND CLADOGRAMS ARE MODELS OF EVOLUTIONARY HISTORY THAT CAN BE TESTED Phylogeny is the history of descent of organisms from their common ancestor. Phylogenetic
CLADISTICS Student Packet SUMMARY PHYLOGENETIC TREES AND CLADOGRAMS ARE MODELS OF EVOLUTIONARY HISTORY THAT CAN BE TESTED Phylogeny is the history of descent of organisms from their common ancestor. Phylogenetic
Phelsuma ISSN Volume 9 (supplement A) 2001
 Phelsuma ISSN 1026-5023 Volume 9 (supplement A) 2001 Tortoise phylogeny and the Geochelone problem J. Gerlach 133 Cherry Hinton Road, Cambridge CB1 7BX, UK PO Box 207, Victoria, Mahe, SEYCHELLES jstgerlach@aol.com
Phelsuma ISSN 1026-5023 Volume 9 (supplement A) 2001 Tortoise phylogeny and the Geochelone problem J. Gerlach 133 Cherry Hinton Road, Cambridge CB1 7BX, UK PO Box 207, Victoria, Mahe, SEYCHELLES jstgerlach@aol.com
Bio 1B Lecture Outline (please print and bring along) Fall, 2006
 Bio 1B Lecture Outline (please print and bring along) Fall, 2006 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #4 -- Phylogenetic Analysis (Cladistics) -- Oct.
Bio 1B Lecture Outline (please print and bring along) Fall, 2006 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #4 -- Phylogenetic Analysis (Cladistics) -- Oct.
Phylogeny Reconstruction
 Phylogeny Reconstruction Trees, Methods and Characters Reading: Gregory, 2008. Understanding Evolutionary Trees (Polly, 2006) Lab tomorrow Meet in Geology GY522 Bring computers if you have them (they will
Phylogeny Reconstruction Trees, Methods and Characters Reading: Gregory, 2008. Understanding Evolutionary Trees (Polly, 2006) Lab tomorrow Meet in Geology GY522 Bring computers if you have them (they will
Phylogenetic hypotheses for the turtle family Geoemydidae q
 Molecular Phylogenetics and Evolution 32 (2004) 164 182 MOLECULAR PHYLOGENETICS AND EVOLUTION www.elsevier.com/locate/ympev Phylogenetic hypotheses for the turtle family Geoemydidae q Phillip Q. Spinks,
Molecular Phylogenetics and Evolution 32 (2004) 164 182 MOLECULAR PHYLOGENETICS AND EVOLUTION www.elsevier.com/locate/ympev Phylogenetic hypotheses for the turtle family Geoemydidae q Phillip Q. Spinks,
Cladistics (reading and making of cladograms)
 Cladistics (reading and making of cladograms) Definitions Systematics The branch of biological sciences concerned with classifying organisms Taxon (pl: taxa) Any unit of biological diversity (eg. Animalia,
Cladistics (reading and making of cladograms) Definitions Systematics The branch of biological sciences concerned with classifying organisms Taxon (pl: taxa) Any unit of biological diversity (eg. Animalia,
A Mitochondrial DNA Phylogeny of Extant Species of the Genus Trachemys with Resulting Taxonomic Implications
 NOTES AND FIELD REPORTS 131 Chelonian Conservation and Biology, 2008, 7(1): 131 135 Ó 2008 Chelonian Research Foundation A Mitochondrial DNA Phylogeny of Extant Species of the Genus Trachemys with Resulting
NOTES AND FIELD REPORTS 131 Chelonian Conservation and Biology, 2008, 7(1): 131 135 Ó 2008 Chelonian Research Foundation A Mitochondrial DNA Phylogeny of Extant Species of the Genus Trachemys with Resulting
Title: Phylogenetic Methods and Vertebrate Phylogeny
 Title: Phylogenetic Methods and Vertebrate Phylogeny Central Question: How can evolutionary relationships be determined objectively? Sub-questions: 1. What affect does the selection of the outgroup have
Title: Phylogenetic Methods and Vertebrate Phylogeny Central Question: How can evolutionary relationships be determined objectively? Sub-questions: 1. What affect does the selection of the outgroup have
UNIT III A. Descent with Modification(Ch19) B. Phylogeny (Ch20) C. Evolution of Populations (Ch21) D. Origin of Species or Speciation (Ch22)
 UNIT III A. Descent with Modification(Ch9) B. Phylogeny (Ch2) C. Evolution of Populations (Ch2) D. Origin of Species or Speciation (Ch22) Classification in broad term simply means putting things in classes
UNIT III A. Descent with Modification(Ch9) B. Phylogeny (Ch2) C. Evolution of Populations (Ch2) D. Origin of Species or Speciation (Ch22) Classification in broad term simply means putting things in classes
Red - Footed Tortoises In Captivity (With Notes On Yellow - Footed Tortoises By Amanda Ebenhack READ ONLINE
 Red - Footed Tortoises In Captivity (With Notes On Yellow - Footed Tortoises By Amanda Ebenhack READ ONLINE Exportation for the pet trade also has a negative effect on yellow-footed tortoises In captivity,
Red - Footed Tortoises In Captivity (With Notes On Yellow - Footed Tortoises By Amanda Ebenhack READ ONLINE Exportation for the pet trade also has a negative effect on yellow-footed tortoises In captivity,
History of Lineages. Chapter 11. Jamie Oaks 1. April 11, Kincaid Hall 524. c 2007 Boris Kulikov boris-kulikov.blogspot.
 History of Lineages Chapter 11 Jamie Oaks 1 1 Kincaid Hall 524 joaks1@gmail.com April 11, 2014 c 2007 Boris Kulikov boris-kulikov.blogspot.com History of Lineages J. Oaks, University of Washington 1/46
History of Lineages Chapter 11 Jamie Oaks 1 1 Kincaid Hall 524 joaks1@gmail.com April 11, 2014 c 2007 Boris Kulikov boris-kulikov.blogspot.com History of Lineages J. Oaks, University of Washington 1/46
Fig Phylogeny & Systematics
 Fig. 26- Phylogeny & Systematics Tree of Life phylogenetic relationship for 3 clades (http://evolution.berkeley.edu Fig. 26-2 Phylogenetic tree Figure 26.3 Taxonomy Taxon Carolus Linnaeus Species: Panthera
Fig. 26- Phylogeny & Systematics Tree of Life phylogenetic relationship for 3 clades (http://evolution.berkeley.edu Fig. 26-2 Phylogenetic tree Figure 26.3 Taxonomy Taxon Carolus Linnaeus Species: Panthera
Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: Evidence for a misleading mitochondrial gene tree
 Molecular Phylogenetics and Evolution 39 (2006) 628 644 www.elsevier.com/locate/ympev Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: Evidence for a misleading
Molecular Phylogenetics and Evolution 39 (2006) 628 644 www.elsevier.com/locate/ympev Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: Evidence for a misleading
INQUIRY & INVESTIGATION
 INQUIRY & INVESTIGTION Phylogenies & Tree-Thinking D VID. UM SUSN OFFNER character a trait or feature that varies among a set of taxa (e.g., hair color) character-state a variant of a character that occurs
INQUIRY & INVESTIGTION Phylogenies & Tree-Thinking D VID. UM SUSN OFFNER character a trait or feature that varies among a set of taxa (e.g., hair color) character-state a variant of a character that occurs
Modern Evolutionary Classification. Lesson Overview. Lesson Overview Modern Evolutionary Classification
 Lesson Overview 18.2 Modern Evolutionary Classification THINK ABOUT IT Darwin s ideas about a tree of life suggested a new way to classify organisms not just based on similarities and differences, but
Lesson Overview 18.2 Modern Evolutionary Classification THINK ABOUT IT Darwin s ideas about a tree of life suggested a new way to classify organisms not just based on similarities and differences, but
The evolutionary origin of Indian Ocean tortoises (Dipsochelys)
 Molecular Phylogenetics and Evolution 24 (2002) 216 227 MOLECULAR PHYLOGENETICS AND EVOLUTION www.academicpress.com The evolutionary origin of Indian Ocean tortoises (Dipsochelys) Eric P. Palkovacs, a,
Molecular Phylogenetics and Evolution 24 (2002) 216 227 MOLECULAR PHYLOGENETICS AND EVOLUTION www.academicpress.com The evolutionary origin of Indian Ocean tortoises (Dipsochelys) Eric P. Palkovacs, a,
1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters
 1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. The sister group of J. K b. The sister group
1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. The sister group of J. K b. The sister group
muscles (enhancing biting strength). Possible states: none, one, or two.
 Reconstructing Evolutionary Relationships S-1 Practice Exercise: Phylogeny of Terrestrial Vertebrates In this example we will construct a phylogenetic hypothesis of the relationships between seven taxa
Reconstructing Evolutionary Relationships S-1 Practice Exercise: Phylogeny of Terrestrial Vertebrates In this example we will construct a phylogenetic hypothesis of the relationships between seven taxa
Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1
 Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1 Systematics is the comparative study of biological diversity with the intent of determining the relationships between organisms. Humankind has always
Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1 Systematics is the comparative study of biological diversity with the intent of determining the relationships between organisms. Humankind has always
Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes)
 Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes) Phylogenetics is the study of the relationships of organisms to each other.
Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes) Phylogenetics is the study of the relationships of organisms to each other.
Introduction to Cladistic Analysis
 3.0 Copyright 2008 by Department of Integrative Biology, University of California-Berkeley Introduction to Cladistic Analysis tunicate lamprey Cladoselache trout lungfish frog four jaws swimbladder or
3.0 Copyright 2008 by Department of Integrative Biology, University of California-Berkeley Introduction to Cladistic Analysis tunicate lamprey Cladoselache trout lungfish frog four jaws swimbladder or
Release of Arnold s giant tortoises Dipsochelys arnoldi on Silhouette island, Seychelles
 Release of Arnold s giant tortoises Dipsochelys arnoldi on Silhouette island, Seychelles Justin Gerlach Nature Protection Trust of Seychelles jstgerlach@aol.com Summary On 7 th December 2007 five adult
Release of Arnold s giant tortoises Dipsochelys arnoldi on Silhouette island, Seychelles Justin Gerlach Nature Protection Trust of Seychelles jstgerlach@aol.com Summary On 7 th December 2007 five adult
LABORATORY EXERCISE 6: CLADISTICS I
 Biology 4415/5415 Evolution LABORATORY EXERCISE 6: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Biology 4415/5415 Evolution LABORATORY EXERCISE 6: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Systematics, Taxonomy and Conservation. Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem
 Systematics, Taxonomy and Conservation Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem What is expected of you? Part I: develop and print the cladogram there
Systematics, Taxonomy and Conservation Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem What is expected of you? Part I: develop and print the cladogram there
HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS
 HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS WASHINGTON AND LONDON 995 by the Smithsonian Institution All rights reserved
HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS WASHINGTON AND LONDON 995 by the Smithsonian Institution All rights reserved
Evolution of Agamidae. species spanning Asia, Africa, and Australia. Archeological specimens and other data
 Evolution of Agamidae Jeff Blackburn Biology 303 Term Paper 11-14-2003 Agamidae is a family of squamates, including 53 genera and over 300 extant species spanning Asia, Africa, and Australia. Archeological
Evolution of Agamidae Jeff Blackburn Biology 303 Term Paper 11-14-2003 Agamidae is a family of squamates, including 53 genera and over 300 extant species spanning Asia, Africa, and Australia. Archeological
Inferring Ancestor-Descendant Relationships in the Fossil Record
 Inferring Ancestor-Descendant Relationships in the Fossil Record (With Statistics) David Bapst, Melanie Hopkins, April Wright, Nick Matzke & Graeme Lloyd GSA 2016 T151 Wednesday Sept 28 th, 9:15 AM Feel
Inferring Ancestor-Descendant Relationships in the Fossil Record (With Statistics) David Bapst, Melanie Hopkins, April Wright, Nick Matzke & Graeme Lloyd GSA 2016 T151 Wednesday Sept 28 th, 9:15 AM Feel
These small issues are easily addressed by small changes in wording, and should in no way delay publication of this first- rate paper.
 Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper reports on a highly significant discovery and associated analysis that are likely to be of broad interest to the scientific community.
Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper reports on a highly significant discovery and associated analysis that are likely to be of broad interest to the scientific community.
LABORATORY EXERCISE 7: CLADISTICS I
 Biology 4415/5415 Evolution LABORATORY EXERCISE 7: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Biology 4415/5415 Evolution LABORATORY EXERCISE 7: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Description of an anomalous tortoise (Reptilia: Testudinidae) from the Early Holocene of Zimbabwe
 Description of an anomalous tortoise (Reptilia: Testudinidae) from the Early Holocene of Zimbabwe Donald G. Broadley* Research Associate, Natural History Museum of Zimbabwe, Bulawayo, Zimbabwe Received
Description of an anomalous tortoise (Reptilia: Testudinidae) from the Early Holocene of Zimbabwe Donald G. Broadley* Research Associate, Natural History Museum of Zimbabwe, Bulawayo, Zimbabwe Received
Phylogeographic assessment of Acanthodactylus boskianus (Reptilia: Lacertidae) based on phylogenetic analysis of mitochondrial DNA.
 Zoology Department Phylogeographic assessment of Acanthodactylus boskianus (Reptilia: Lacertidae) based on phylogenetic analysis of mitochondrial DNA By HAGAR IBRAHIM HOSNI BAYOUMI A thesis submitted in
Zoology Department Phylogeographic assessment of Acanthodactylus boskianus (Reptilia: Lacertidae) based on phylogenetic analysis of mitochondrial DNA By HAGAR IBRAHIM HOSNI BAYOUMI A thesis submitted in
DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS
 Syst. Biol. 45(4):39^14, 1996 DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS STEVEN POE Department of Zoology and Texas Memorial Museum, University of Texas, Austin, Texas 78712-1064, USA; E-mail:
Syst. Biol. 45(4):39^14, 1996 DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS STEVEN POE Department of Zoology and Texas Memorial Museum, University of Texas, Austin, Texas 78712-1064, USA; E-mail:
17.2 Classification Based on Evolutionary Relationships Organization of all that speciation!
 Organization of all that speciation! Patterns of evolution.. Taxonomy gets an over haul! Using more than morphology! 3 domains, 6 kingdoms KEY CONCEPT Modern classification is based on evolutionary relationships.
Organization of all that speciation! Patterns of evolution.. Taxonomy gets an over haul! Using more than morphology! 3 domains, 6 kingdoms KEY CONCEPT Modern classification is based on evolutionary relationships.
What are taxonomy, classification, and systematics?
 Topic 2: Comparative Method o Taxonomy, classification, systematics o Importance of phylogenies o A closer look at systematics o Some key concepts o Parts of a cladogram o Groups and characters o Homology
Topic 2: Comparative Method o Taxonomy, classification, systematics o Importance of phylogenies o A closer look at systematics o Some key concepts o Parts of a cladogram o Groups and characters o Homology
6. The lifetime Darwinian fitness of one organism is greater than that of another organism if: A. it lives longer than the other B. it is able to outc
 1. The money in the kingdom of Florin consists of bills with the value written on the front, and pictures of members of the royal family on the back. To test the hypothesis that all of the Florinese $5
1. The money in the kingdom of Florin consists of bills with the value written on the front, and pictures of members of the royal family on the back. To test the hypothesis that all of the Florinese $5
Interspecific hybridization between Mauremys reevesii and Mauremys sinensis: Evidence from morphology and DNA sequence data
 African Journal of Biotechnology Vol. 10(35), pp. 6716-6724, 13 July, 2011 Available online at http://www.academicjournals.org/ajb DOI: 10.5897/AJB11.063 ISSN 1684 5315 2011 Academic Journals Full Length
African Journal of Biotechnology Vol. 10(35), pp. 6716-6724, 13 July, 2011 Available online at http://www.academicjournals.org/ajb DOI: 10.5897/AJB11.063 ISSN 1684 5315 2011 Academic Journals Full Length
Molecular Phylogenetics and Evolution
 Molecular Phylogenetics and Evolution 59 (2011) 623 635 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev A multigenic perspective
Molecular Phylogenetics and Evolution 59 (2011) 623 635 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev A multigenic perspective
1 Describe the anatomy and function of the turtle shell. 2 Describe respiration in turtles. How does the shell affect respiration?
 GVZ 2017 Practice Questions Set 1 Test 3 1 Describe the anatomy and function of the turtle shell. 2 Describe respiration in turtles. How does the shell affect respiration? 3 According to the most recent
GVZ 2017 Practice Questions Set 1 Test 3 1 Describe the anatomy and function of the turtle shell. 2 Describe respiration in turtles. How does the shell affect respiration? 3 According to the most recent
Centre of Macaronesian Studies, University of Madeira, Penteada, 9000 Funchal, Portugal b
 Molecular Phylogenetics and Evolution 34 (2005) 480 485 www.elsevier.com/locate/ympev Phylogenetic relationships of Hemidactylus geckos from the Gulf of Guinea islands: patterns of natural colonizations
Molecular Phylogenetics and Evolution 34 (2005) 480 485 www.elsevier.com/locate/ympev Phylogenetic relationships of Hemidactylus geckos from the Gulf of Guinea islands: patterns of natural colonizations
Horned lizard (Phrynosoma) phylogeny inferred from mitochondrial genes and morphological characters: understanding conflicts using multiple approaches
 Molecular Phylogenetics and Evolution xxx (2004) xxx xxx MOLECULAR PHYLOGENETICS AND EVOLUTION www.elsevier.com/locate/ympev Horned lizard (Phrynosoma) phylogeny inferred from mitochondrial genes and morphological
Molecular Phylogenetics and Evolution xxx (2004) xxx xxx MOLECULAR PHYLOGENETICS AND EVOLUTION www.elsevier.com/locate/ympev Horned lizard (Phrynosoma) phylogeny inferred from mitochondrial genes and morphological
Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes)
 Molecular Phylogenetics and Evolution 39 (2006) 91 110 www.elsevier.com/locate/ympev Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes) Todd A. Castoe, Christopher L. Parkinson
Molecular Phylogenetics and Evolution 39 (2006) 91 110 www.elsevier.com/locate/ympev Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes) Todd A. Castoe, Christopher L. Parkinson
Validity of Pelodiscus parviformis (Testudines: Trionychidae) Inferred from Molecular and Morphological Analyses
 Asian Herpetological Research 2011, 2(1): 21-29 DOI: 10.3724/SP.J.1245.2011.00021 Validity of Pelodiscus parviformis (Testudines: Trionychidae) Inferred from Molecular and Morphological Analyses Ping YANG,
Asian Herpetological Research 2011, 2(1): 21-29 DOI: 10.3724/SP.J.1245.2011.00021 Validity of Pelodiscus parviformis (Testudines: Trionychidae) Inferred from Molecular and Morphological Analyses Ping YANG,
Ch 1.2 Determining How Species Are Related.notebook February 06, 2018
 Name 3 "Big Ideas" from our last notebook lecture: * * * 1 WDYR? Of the following organisms, which is the closest relative of the "Snowy Owl" (Bubo scandiacus)? a) barn owl (Tyto alba) b) saw whet owl
Name 3 "Big Ideas" from our last notebook lecture: * * * 1 WDYR? Of the following organisms, which is the closest relative of the "Snowy Owl" (Bubo scandiacus)? a) barn owl (Tyto alba) b) saw whet owl
APPLICATION OF BODY CONDITION INDICES FOR LEOPARD TORTOISES (GEOCHELONE PARDALIS)
 APPLICATION OF BODY CONDITION INDICES FOR LEOPARD TORTOISES (GEOCHELONE PARDALIS) Laura Lickel, BS,* and Mark S. Edwards, Ph. California Polytechnic State University, Animal Science Department, San Luis
APPLICATION OF BODY CONDITION INDICES FOR LEOPARD TORTOISES (GEOCHELONE PARDALIS) Laura Lickel, BS,* and Mark S. Edwards, Ph. California Polytechnic State University, Animal Science Department, San Luis
A phylogeny for side-necked turtles (Chelonia: Pleurodira) based on mitochondrial and nuclear gene sequence variation
 Bivlogkal Journal ofthe Linnean So&& (1998), 67: 2 13-246. \\'ith 4 figures Article ID biji.1998.0300, avaiiable online at http://www.idealihrary.lom on IDE kt @ c A phylogeny for side-necked turtles (Chelonia:
Bivlogkal Journal ofthe Linnean So&& (1998), 67: 2 13-246. \\'ith 4 figures Article ID biji.1998.0300, avaiiable online at http://www.idealihrary.lom on IDE kt @ c A phylogeny for side-necked turtles (Chelonia:
GEODIS 2.0 DOCUMENTATION
 GEODIS.0 DOCUMENTATION 1999-000 David Posada and Alan Templeton Contact: David Posada, Department of Zoology, 574 WIDB, Provo, UT 8460-555, USA Fax: (801) 78 74 e-mail: dp47@email.byu.edu 1. INTRODUCTION
GEODIS.0 DOCUMENTATION 1999-000 David Posada and Alan Templeton Contact: David Posada, Department of Zoology, 574 WIDB, Provo, UT 8460-555, USA Fax: (801) 78 74 e-mail: dp47@email.byu.edu 1. INTRODUCTION
1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters
 1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. Identify the taxon (or taxa if there is more
1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. Identify the taxon (or taxa if there is more
Sparse Supermatrices for Phylogenetic Inference: Taxonomy, Alignment, Rogue Taxa, and the Phylogeny of Living Turtles
 Syst. Biol. 59(1):42 58, 2010 c The Author(s) 2009. Published by Oxford University Press, on behalf of the Society of Systematic Biologists. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Syst. Biol. 59(1):42 58, 2010 c The Author(s) 2009. Published by Oxford University Press, on behalf of the Society of Systematic Biologists. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Systematics and taxonomy of the genus Culicoides what is coming next?
 Systematics and taxonomy of the genus Culicoides what is coming next? Claire Garros 1, Bruno Mathieu 2, Thomas Balenghien 1, Jean-Claude Delécolle 2 1 CIRAD, Montpellier, France 2 IPPTS, Strasbourg, France
Systematics and taxonomy of the genus Culicoides what is coming next? Claire Garros 1, Bruno Mathieu 2, Thomas Balenghien 1, Jean-Claude Delécolle 2 1 CIRAD, Montpellier, France 2 IPPTS, Strasbourg, France
PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, NY 10024
 PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, NY 10024 Number 3365, 61 pp., 7 figures, 3 tables May 17, 2002 Phylogenetic Relationships of Whiptail Lizards
PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, NY 10024 Number 3365, 61 pp., 7 figures, 3 tables May 17, 2002 Phylogenetic Relationships of Whiptail Lizards
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution
 Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
Biodiversity and Distributions. Lecture 2: Biodiversity. The process of natural selection
 Lecture 2: Biodiversity What is biological diversity? Natural selection Adaptive radiations and convergent evolution Biogeography Biodiversity and Distributions Types of biological diversity: Genetic diversity
Lecture 2: Biodiversity What is biological diversity? Natural selection Adaptive radiations and convergent evolution Biogeography Biodiversity and Distributions Types of biological diversity: Genetic diversity
No limbs Eastern glass lizard. Monitor lizard. Iguanas. ANCESTRAL LIZARD (with limbs) Snakes. No limbs. Geckos Pearson Education, Inc.
 No limbs Eastern glass lizard Monitor lizard guanas ANCESTRAL LZARD (with limbs) No limbs Snakes Geckos Species: Panthera pardus Genus: Panthera Family: Felidae Order: Carnivora Class: Mammalia Phylum:
No limbs Eastern glass lizard Monitor lizard guanas ANCESTRAL LZARD (with limbs) No limbs Snakes Geckos Species: Panthera pardus Genus: Panthera Family: Felidae Order: Carnivora Class: Mammalia Phylum:
Biodiversity and Extinction. Lecture 9
 Biodiversity and Extinction Lecture 9 This lecture will help you understand: The scope of Earth s biodiversity Levels and patterns of biodiversity Mass extinction vs background extinction Attributes of
Biodiversity and Extinction Lecture 9 This lecture will help you understand: The scope of Earth s biodiversity Levels and patterns of biodiversity Mass extinction vs background extinction Attributes of
Modern taxonomy. Building family trees 10/10/2011. Knowing a lot about lots of creatures. Tom Hartman. Systematics includes: 1.
 Modern taxonomy Building family trees Tom Hartman www.tuatara9.co.uk Classification has moved away from the simple grouping of organisms according to their similarities (phenetics) and has become the study
Modern taxonomy Building family trees Tom Hartman www.tuatara9.co.uk Classification has moved away from the simple grouping of organisms according to their similarities (phenetics) and has become the study
Molecular Systematics and Evolution of Regina and the Thamnophiine Snakes
 Molecular Phylogenetics and Evolution Vol. 21, No. 3, December, pp. 408 423, 2001 doi:10.1006/mpev.2001.1024, available online at http://www.idealibrary.com on Molecular Systematics and Evolution of Regina
Molecular Phylogenetics and Evolution Vol. 21, No. 3, December, pp. 408 423, 2001 doi:10.1006/mpev.2001.1024, available online at http://www.idealibrary.com on Molecular Systematics and Evolution of Regina
May 10, SWBAT analyze and evaluate the scientific evidence provided by the fossil record.
 May 10, 2017 Aims: SWBAT analyze and evaluate the scientific evidence provided by the fossil record. Agenda 1. Do Now 2. Class Notes 3. Guided Practice 4. Independent Practice 5. Practicing our AIMS: E.3-Examining
May 10, 2017 Aims: SWBAT analyze and evaluate the scientific evidence provided by the fossil record. Agenda 1. Do Now 2. Class Notes 3. Guided Practice 4. Independent Practice 5. Practicing our AIMS: E.3-Examining
of Veterinary and Pharmaceutical Sciences Brno, Palackeho tr. 1/3, Brno, , Czech Republic
 Biological Journal of the Linnean Society, 2016, 117, 305 321. Comparative phylogeographies of six species of hinged terrapins (Pelusios spp.) reveal discordant patterns and unexpected differentiation
Biological Journal of the Linnean Society, 2016, 117, 305 321. Comparative phylogeographies of six species of hinged terrapins (Pelusios spp.) reveal discordant patterns and unexpected differentiation
Multiple Data Sets, High Homoplasy, and the Phylogeny of Softshell Turtles (Testudines: Trionychidae)
 Syst. Biol. 53(5):693 710, 2004 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150490503053 Multiple Data Sets, High Homoplasy, and the Phylogeny
Syst. Biol. 53(5):693 710, 2004 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150490503053 Multiple Data Sets, High Homoplasy, and the Phylogeny
Caecilians (Gymnophiona)
 Caecilians (Gymnophiona) David J. Gower* and Mark Wilkinson Department of Zoology, The Natural History Museum, London SW7 5BD, UK *To whom correspondence should be addressed (d.gower@nhm. ac.uk) Abstract
Caecilians (Gymnophiona) David J. Gower* and Mark Wilkinson Department of Zoology, The Natural History Museum, London SW7 5BD, UK *To whom correspondence should be addressed (d.gower@nhm. ac.uk) Abstract
Phylogenetic diversity of endangered and critically endangered southeast Asian softshell turtles (Trionychidae: Chitra)
 Biological Conservation 104 (2002) 173 179 www.elsevier.com/locate/biocon Phylogenetic diversity of endangered and critically endangered southeast Asian softshell turtles (Trionychidae: Chitra) Tag N.
Biological Conservation 104 (2002) 173 179 www.elsevier.com/locate/biocon Phylogenetic diversity of endangered and critically endangered southeast Asian softshell turtles (Trionychidae: Chitra) Tag N.
SUPPLEMENTARY INFORMATION
 In comparison to Proganochelys (Gaffney, 1990), Odontochelys semitestacea is a small turtle. The adult status of the specimen is documented not only by the generally well-ossified appendicular skeleton
In comparison to Proganochelys (Gaffney, 1990), Odontochelys semitestacea is a small turtle. The adult status of the specimen is documented not only by the generally well-ossified appendicular skeleton
A NOMENCLATURAL HISTORY OF TORTOISES
 A NOMENCLATURAL HISTORY OF TORTOISES (Family Testudinidae) Charles R Crumly Department of Herpetology San Diego Naturtal History Museum SMITHSONIAN HERPETOLOGICAL INFORMATION SERVICE NO 75 1988 SMITHSONIAN
A NOMENCLATURAL HISTORY OF TORTOISES (Family Testudinidae) Charles R Crumly Department of Herpetology San Diego Naturtal History Museum SMITHSONIAN HERPETOLOGICAL INFORMATION SERVICE NO 75 1988 SMITHSONIAN
Testing Phylogenetic Hypotheses with Molecular Data 1
 Testing Phylogenetic Hypotheses with Molecular Data 1 How does an evolutionary biologist quantify the timing and pathways for diversification (speciation)? If we observe diversification today, the processes
Testing Phylogenetic Hypotheses with Molecular Data 1 How does an evolutionary biologist quantify the timing and pathways for diversification (speciation)? If we observe diversification today, the processes
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST In this laboratory investigation, you will use BLAST to compare several genes, and then use the information to construct a cladogram.
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST In this laboratory investigation, you will use BLAST to compare several genes, and then use the information to construct a cladogram.
Prof. Neil. J.L. Heideman
 Prof. Neil. J.L. Heideman Position Office Mailing address E-mail : Vice-dean (Professor of Zoology) : No. 10, Biology Building : P.O. Box 339 (Internal Box 44), Bloemfontein 9300, South Africa : heidemannj.sci@mail.uovs.ac.za
Prof. Neil. J.L. Heideman Position Office Mailing address E-mail : Vice-dean (Professor of Zoology) : No. 10, Biology Building : P.O. Box 339 (Internal Box 44), Bloemfontein 9300, South Africa : heidemannj.sci@mail.uovs.ac.za
Required and Recommended Supporting Information for IUCN Red List Assessments
 Required and Recommended Supporting Information for IUCN Red List Assessments This is Annex 1 of the Rules of Procedure for IUCN Red List Assessments 2017 2020 as approved by the IUCN SSC Steering Committee
Required and Recommended Supporting Information for IUCN Red List Assessments This is Annex 1 of the Rules of Procedure for IUCN Red List Assessments 2017 2020 as approved by the IUCN SSC Steering Committee
Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per.
 Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per. Introduction Imagine a single diagram representing the evolutionary relationships between everything that has ever lived. If life evolved
Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per. Introduction Imagine a single diagram representing the evolutionary relationships between everything that has ever lived. If life evolved
Evolution as Fact. The figure below shows transitional fossils in the whale lineage.
 Evolution as Fact Evolution is a fact. Organisms descend from others with modification. Phylogeny, the lineage of ancestors and descendants, is the scientific term to Darwin's phrase "descent with modification."
Evolution as Fact Evolution is a fact. Organisms descend from others with modification. Phylogeny, the lineage of ancestors and descendants, is the scientific term to Darwin's phrase "descent with modification."
The Making of the Fittest: LESSON STUDENT MATERIALS USING DNA TO EXPLORE LIZARD PHYLOGENY
 The Making of the Fittest: Natural The The Making Origin Selection of the of Species and Fittest: Adaptation Natural Lizards Selection in an Evolutionary and Adaptation Tree INTRODUCTION USING DNA TO EXPLORE
The Making of the Fittest: Natural The The Making Origin Selection of the of Species and Fittest: Adaptation Natural Lizards Selection in an Evolutionary and Adaptation Tree INTRODUCTION USING DNA TO EXPLORE
Proopiomelanocortin (POMC) and testing the phylogenetic position of turtles (Testudines)
 Accepted on 10 November 2010 J Zool Syst Evol Res Department of Biological Sciences, Southeastern Louisiana University, Hammond, LA, USA Proopiomelanocortin (POMC) and testing the phylogenetic position
Accepted on 10 November 2010 J Zool Syst Evol Res Department of Biological Sciences, Southeastern Louisiana University, Hammond, LA, USA Proopiomelanocortin (POMC) and testing the phylogenetic position
HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION
 Syst. Zool., 3(3), 98, pp. 229-249 HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION DANIEL R. BROOKS Abstract Brooks, ID. R. (Department of Zoology, University of British Columbia, 275 Wesbrook Mall,
Syst. Zool., 3(3), 98, pp. 229-249 HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION DANIEL R. BROOKS Abstract Brooks, ID. R. (Department of Zoology, University of British Columbia, 275 Wesbrook Mall,
Warm-Up: Fill in the Blank
 Warm-Up: Fill in the Blank 1. For natural selection to happen, there must be variation in the population. 2. The preserved remains of organisms, called provides evidence for evolution. 3. By using and
Warm-Up: Fill in the Blank 1. For natural selection to happen, there must be variation in the population. 2. The preserved remains of organisms, called provides evidence for evolution. 3. By using and
Phylogeny of snakes (Serpentes): combining morphological and molecular data in likelihood, Bayesian and parsimony analyses
 Systematics and Biodiversity 5 (4): 371 389 Issued 20 November 2007 doi:10.1017/s1477200007002290 Printed in the United Kingdom C The Natural History Museum Phylogeny of snakes (Serpentes): combining morphological
Systematics and Biodiversity 5 (4): 371 389 Issued 20 November 2007 doi:10.1017/s1477200007002290 Printed in the United Kingdom C The Natural History Museum Phylogeny of snakes (Serpentes): combining morphological
8/19/2013. Topic 4: The Origin of Tetrapods. Topic 4: The Origin of Tetrapods. The geological time scale. The geological time scale.
 Topic 4: The Origin of Tetrapods Next two lectures will deal with: Origin of Tetrapods, transition from water to land. Origin of Amniotes, transition to dry habitats. Topic 4: The Origin of Tetrapods What
Topic 4: The Origin of Tetrapods Next two lectures will deal with: Origin of Tetrapods, transition from water to land. Origin of Amniotes, transition to dry habitats. Topic 4: The Origin of Tetrapods What
The phylogeny of antiarch placoderms. Sarah Kearsley Geology 394 Senior Thesis
 The phylogeny of antiarch placoderms Sarah Kearsley Geology 394 Senior Thesis Abstract The most comprehensive phylogenetic study of antiarchs to date (Zhu, 1996) included information not derived from observation.
The phylogeny of antiarch placoderms Sarah Kearsley Geology 394 Senior Thesis Abstract The most comprehensive phylogenetic study of antiarchs to date (Zhu, 1996) included information not derived from observation.
ADAPTIVE EVOLUTION OF PLASTRON SHAPE IN EMYDINE TURTLES
 ORIGINAL ARTICLE doi:10.1111/j.1558-5646.2010.01118.x ADAPTIVE EVOLUTION OF PLASTRON SHAPE IN EMYDINE TURTLES Kenneth D. Angielczyk, 1,2 Chris R. Feldman, 3,4 and Gretchen R. Miller 5,6 1 Department of
ORIGINAL ARTICLE doi:10.1111/j.1558-5646.2010.01118.x ADAPTIVE EVOLUTION OF PLASTRON SHAPE IN EMYDINE TURTLES Kenneth D. Angielczyk, 1,2 Chris R. Feldman, 3,4 and Gretchen R. Miller 5,6 1 Department of
Do the traits of organisms provide evidence for evolution?
 PhyloStrat Tutorial Do the traits of organisms provide evidence for evolution? Consider two hypotheses about where Earth s organisms came from. The first hypothesis is from John Ray, an influential British
PhyloStrat Tutorial Do the traits of organisms provide evidence for evolution? Consider two hypotheses about where Earth s organisms came from. The first hypothesis is from John Ray, an influential British
Was there a second adaptive radiation of giant tortoises in
 Molecular Ecology (2003) 12, 1415 1424 doi: 10.1046/j.1365-294X.2003.01842.x Was there a second adaptive radiation of giant tortoises in Blackwell Publishing Ltd. the Indian Ocean? Using mitochondrial
Molecular Ecology (2003) 12, 1415 1424 doi: 10.1046/j.1365-294X.2003.01842.x Was there a second adaptive radiation of giant tortoises in Blackwell Publishing Ltd. the Indian Ocean? Using mitochondrial
Molecular Phylogenetics and Evolution
 Molecular Phylogenetics and Evolution 49 (2008) 92 101 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev The genus Coleodactylus
Molecular Phylogenetics and Evolution 49 (2008) 92 101 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev The genus Coleodactylus
Are Turtles Diapsid Reptiles?
 Are Turtles Diapsid Reptiles? Jack K. Horner P.O. Box 266 Los Alamos NM 87544 USA BIOCOMP 2013 Abstract It has been argued that, based on a neighbor-joining analysis of a broad set of fossil reptile morphological
Are Turtles Diapsid Reptiles? Jack K. Horner P.O. Box 266 Los Alamos NM 87544 USA BIOCOMP 2013 Abstract It has been argued that, based on a neighbor-joining analysis of a broad set of fossil reptile morphological
The impact of the recognizing evolution on systematics
 The impact of the recognizing evolution on systematics 1. Genealogical relationships between species could serve as the basis for taxonomy 2. Two sources of similarity: (a) similarity from descent (b)
The impact of the recognizing evolution on systematics 1. Genealogical relationships between species could serve as the basis for taxonomy 2. Two sources of similarity: (a) similarity from descent (b)
Sample Questions: EXAMINATION I Form A Mammalogy -EEOB 625. Name Composite of previous Examinations
 Sample Questions: EXAMINATION I Form A Mammalogy -EEOB 625 Name Composite of previous Examinations Part I. Define or describe only 5 of the following 6 words - 15 points (3 each). If you define all 6,
Sample Questions: EXAMINATION I Form A Mammalogy -EEOB 625 Name Composite of previous Examinations Part I. Define or describe only 5 of the following 6 words - 15 points (3 each). If you define all 6,
HONR219D Due 3/29/16 Homework VI
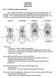 Part 1: Yet More Vertebrate Anatomy!!! HONR219D Due 3/29/16 Homework VI Part 1 builds on homework V by examining the skull in even greater detail. We start with the some of the important bones (thankfully
Part 1: Yet More Vertebrate Anatomy!!! HONR219D Due 3/29/16 Homework VI Part 1 builds on homework V by examining the skull in even greater detail. We start with the some of the important bones (thankfully
Evolution of Biodiversity
 Long term patterns Evolution of Biodiversity Chapter 7 Changes in biodiversity caused by originations and extinctions of taxa over geologic time Analyses of diversity in the fossil record requires procedures
Long term patterns Evolution of Biodiversity Chapter 7 Changes in biodiversity caused by originations and extinctions of taxa over geologic time Analyses of diversity in the fossil record requires procedures
Quiz Flip side of tree creation: EXTINCTION. Knock-on effects (Crooks & Soule, '99)
 Flip side of tree creation: EXTINCTION Quiz 2 1141 1. The Jukes-Cantor model is below. What does the term µt represent? 2. How many ways can you root an unrooted tree with 5 edges? Include a drawing. 3.
Flip side of tree creation: EXTINCTION Quiz 2 1141 1. The Jukes-Cantor model is below. What does the term µt represent? 2. How many ways can you root an unrooted tree with 5 edges? Include a drawing. 3.
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Phylogenetics. Phylogenetic Trees. 1. Represent presumed patterns. 2. Analogous to family trees.
 Phylogenetics. Phylogenetic Trees. 1. Represent presumed patterns of descent. 2. Analogous to family trees. 3. Resolve taxa, e.g., species, into clades each of which includes an ancestral taxon and all
Phylogenetics. Phylogenetic Trees. 1. Represent presumed patterns of descent. 2. Analogous to family trees. 3. Resolve taxa, e.g., species, into clades each of which includes an ancestral taxon and all
Evolution of Birds. Summary:
 Oregon State Standards OR Science 7.1, 7.2, 7.3, 7.3S.1, 7.3S.2 8.1, 8.2, 8.2L.1, 8.3, 8.3S.1, 8.3S.2 H.1, H.2, H.2L.4, H.2L.5, H.3, H.3S.1, H.3S.2, H.3S.3 Summary: Students create phylogenetic trees to
Oregon State Standards OR Science 7.1, 7.2, 7.3, 7.3S.1, 7.3S.2 8.1, 8.2, 8.2L.1, 8.3, 8.3S.1, 8.3S.2 H.1, H.2, H.2L.4, H.2L.5, H.3, H.3S.1, H.3S.2, H.3S.3 Summary: Students create phylogenetic trees to
8/19/2013. Topic 5: The Origin of Amniotes. What are some stem Amniotes? What are some stem Amniotes? The Amniotic Egg. What is an Amniote?
 Topic 5: The Origin of Amniotes Where do amniotes fall out on the vertebrate phylogeny? What are some stem Amniotes? What is an Amniote? What changes were involved with the transition to dry habitats?
Topic 5: The Origin of Amniotes Where do amniotes fall out on the vertebrate phylogeny? What are some stem Amniotes? What is an Amniote? What changes were involved with the transition to dry habitats?
d. Wrist bones. Pacific salmon life cycle. Atlantic salmon (different genus) can spawn more than once.
 Lecture III.5b Answers to HW 1. (2 pts). Tiktaalik bridges the gap between fish and tetrapods by virtue of possessing which of the following? a. Humerus. b. Radius. c. Ulna. d. Wrist bones. 2. (2 pts)
Lecture III.5b Answers to HW 1. (2 pts). Tiktaalik bridges the gap between fish and tetrapods by virtue of possessing which of the following? a. Humerus. b. Radius. c. Ulna. d. Wrist bones. 2. (2 pts)
Comparing DNA Sequence to Understand
 Comparing DNA Sequence to Understand Evolutionary Relationships with BLAST Name: Big Idea 1: Evolution Pre-Reading In order to understand the purposes and learning objectives of this investigation, you
Comparing DNA Sequence to Understand Evolutionary Relationships with BLAST Name: Big Idea 1: Evolution Pre-Reading In order to understand the purposes and learning objectives of this investigation, you
Morphological systematics of kingsnakes, Lampropeltis getula complex (Serpentes: Colubridae), in the eastern United States
 Zootaxa : 1 39 (2006) www.mapress.com/zootaxa/ Copyright 2006 Magnolia Press ISSN 1175-5326 (print edition) ZOOTAXA ISSN 1175-5334 (online edition) Morphological systematics of kingsnakes, Lampropeltis
Zootaxa : 1 39 (2006) www.mapress.com/zootaxa/ Copyright 2006 Magnolia Press ISSN 1175-5326 (print edition) ZOOTAXA ISSN 1175-5334 (online edition) Morphological systematics of kingsnakes, Lampropeltis
Molecular Phylogenetics of Emydine Turtles: Taxonomic Revision and the Evolution of Shell Kinesis
 Molecular Phylogenetics and Evolution Vol. 22, No. 3, March, pp. 388 398, 2002 doi:10.1006/mpev.2001.1070, available online at http://www.idealibrary.com on Molecular Phylogenetics of Emydine Turtles:
Molecular Phylogenetics and Evolution Vol. 22, No. 3, March, pp. 388 398, 2002 doi:10.1006/mpev.2001.1070, available online at http://www.idealibrary.com on Molecular Phylogenetics of Emydine Turtles:
Systematics of the Lizard Family Pygopodidae with Implications for the Diversification of Australian Temperate Biotas
 Syst. Biol. 52(6):757 780, 2003 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150390250974 Systematics of the Lizard Family Pygopodidae with Implications
Syst. Biol. 52(6):757 780, 2003 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150390250974 Systematics of the Lizard Family Pygopodidae with Implications
Phalangeal formulae and ontogenetic variation of carpal morphology in Testudo horsfieldii and T. hermanni
 Amphibia-Reptilia 29 (2008): 93-99 Phalangeal formulae and ontogenetic variation of carpal morphology in Testudo horsfieldii and T. hermanni Ellen Hitschfeld 1, Markus Auer 2, Uwe Fritz 2 Abstract. We
Amphibia-Reptilia 29 (2008): 93-99 Phalangeal formulae and ontogenetic variation of carpal morphology in Testudo horsfieldii and T. hermanni Ellen Hitschfeld 1, Markus Auer 2, Uwe Fritz 2 Abstract. We
