Molecular phylogeny of 48 species of damselfishes (Perciformes: Pomacentridae) using 12S mtdnasequences
|
|
|
- Sophia Charles
- 5 years ago
- Views:
Transcription
1 Molecular Phylogenetics and Evolution 25 (2002) MOLECULAR PHYLOGENETICS AND EVOLUTION Molecular phylogeny of 48 species of damselfishes (Perciformes: Pomacentridae) using 12S mtdnasequences Nian-Hong Jang-Liaw, a Kevin L. Tang, b, * Cho-Fat Hui, c and Kwang-Tsao Shao c a Department of Zoology, National Museum of Natural Science, 1 Kuan-Chien Rd., Tachung, Taiwan 404, ROC b Division of Ichthyology, Natural History Museum and Biodiversity Research Center, Department of Ecology and Evolutionary Biology, Dyche Hall, 1345 Jayhawk Blvd., The University of Kansas, Lawrence, KS , USA c Institute of Zoology, Academia Sinica, Nankang, Taipei 115, Taiwan, ROC Received 18 October 2001; received in revised form 22 April 2002 Abstract Phylogenetic relationships within the family Pomacentridae (Teleostei: Perciformes) were inferred by analyzing a portion of the 12S mitochondrial ribosomal DNAgene. Thirty-four pomacentrid species were sequenced for this study and the resulting data were combined with previously published pomacentrid sequence data to form a combined matrix of 53 pomacentrids representing 48 different species in 18 genera. Four outgroup species were also drawn from published data; these taxa were taken from the other three putative families of the suborder Labroidei, as well as a single representative of the family Moronidae. The data set contained 1053 data columns after alignment according to ribosomal secondary structure and the removal of all ambiguously aligned positions. The resulting strict consensus tree topology generally agreed with the previous molecular hypothesis, and recovers a monophyletic Pomacentridae and subfamily Amphiprioninae. The two other subfamilies included, Chrominae and Pomacentrinae, were found to be polyphyletic. Amonophyletic group consisting of the Amphiprioninae, Pomacentrus, Acanthochromis, Amblyglyphidodon, Neoglyphidodon, Chrysiptera, Neopomacentrus, and Teixeirichthys was found. This group was recovered as the sister group to a clade consisting of a paraphyletic Chromis and a monophyletic Dascyllus. Asister-group relationship between the genus Pomacentrus and the subfamily Amphiprioninae was observed. Ó 2002 Elsevier Science (USA). All rights reserved. 1. Introduction Damselfishes (Teleostei: Pomacentridae) are a diverse and widespread family of primarily marine fishes found throughout the tropical oceans, forming a major component of coral reef communities (Allen, 1975). Although the bulk of damselfish diversity is concentrated in tropical regions, some species are found in temperate waters and others are known from freshwater and brackish environments (Allen, 1989). Pomacentrids are usually small in size, with reef species often no more than 100 mm in length, though several temperate species can get much larger, exceeding 250 mm in standard length (Allen, 1991). Because most species are associated with reefs and tidal zones, damselfishes are usually found near shore and in shallow water (within m * Corresponding author. Fax: address: kltang@ukans.edu (K.L. Tang). of the surface); however, several species do occur at depths greater than 100 m (Allen, 1975). The Pomacentridae is currently recognized as a member of the perciform suborder Labroidei (Nelson, 1994), along with the families Cichlidae, Embiotocidae, and Labridae. However, there is some debate over the monophyly of this suborder. Based on morphological characters, these four families form a monophyletic group (Kaufman and Liem, 1982; Stiassny and Jensen, 1987). However, the monophyly of this suborder has been questioned (Johnson, 1993; Rosen and Patterson, 1990), with a recent molecular study suggesting that the suborder is not monophyletic (Streelman and Karl, 1997). Currently, there are approximately 340 recognized pomacentrid species divided into 29 genera. The family is composed of four subfamilies: Amphiprioninae, Chrominae, Lepidozyginae, and Pomacentrinae (Allen, 1975, 1991). The members of the Amphiprioninae are unique because all of the species have an obligate /02/$ - see front matter Ó 2002 Elsevier Science (USA). All rights reserved. PII: S (02)
2 446 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) symbiotic relationship with sea anemones. The approximately 28 species of this subfamily are united by a number of morphological (Allen, 1972, 1975; Fitzpatrick, 1992; Tang, unpublished) and molecular (Elliott et al., 1999; Tang, 2001) characters. The majority of pomacentrid diversity is concentrated in two of the subfamilies: the Chrominae, with over 80 species, and the Pomacentrinae, with more than 200 species. The Chrominae has been recognized as a natural group on the basis of some morphological characters (Allen, 1975); however, recent molecular analyses have cast some doubt on the monophyly of this subfamily (Tang, 2001). The subfamily Pomacentrinae is not considered a natural assemblage (Allen, 1975), a hypothesis corroborated by DNAdata (Tang, 2001). Species placed in this subfamily are characterized simply as possessing an orbiculate to elongate body shape (Allen, 1975). Finally, Lepidozygus tapeinosoma is the sole representative of the monotypic Lepidozyginae. Prior to Allen (1975), who placed Lepidozygus in its own subfamily, L. tapeinosoma was considered a member of the Chrominae (Norman, 1957). The damselfishes have long posed a challenge to systematists because of their diversity and intraspecific variation (e.g., species can exhibit enormous variation in coloration, both among adults and between juvenile and adult stages). The classification of the Pomacentridae that is in use today was established by Allen (1975, 1991), but, aside from the classification itself, no hypothesis of the relationships within the family was presented. Shao (1986) used morphometric measurements to examine generic relationships within the family. Using morphological characters, Fitzpatrick (1992) found characters supporting the monophyly of the family, but the generic and subfamilial relationships were almost completely unresolved in her analysis. Based on that work and others (Kaufman and Liem, 1982; Lauder and Liem, 1983; Stiassny, 1981), the monophyly of the Pomacentridae is supported by five synapomorphies: (1) a strong sheet of connective tissue originating from the dorsal border of the medial face of the dentary that merges with a cylindrical ligament and inserts onto the ceratohyal (anterior ceratohyal) bone (Stiassny, 1981); (2) a pair of nipple-like processes on the ventral surface of the lower pharyngeal jaw that act as insertion sites for the pharyngohyoideus muscle; (3) a pharyngo-cleithral articulation between the cleithra and the muscular processes of the lower pharyngeal jaw; (4) a prominent obliquus posterior muscle which is separated from the fourth levator externus muscle by a distinct aponeurosis (Kaufman and Liem, 1982; Lauder and Liem, 1983); and (5) the presence of two anal spines (Fitzpatrick, 1992; i.e., first anal pterygiophore with two supernumerary spines and a serially associated soft ray). Tang (2001) used sequence data from the 12S and 16S mitochondrial ribosomal genes to investigate relationships within the family, producing a well-resolved phylogenetic hypothesis of relationships within the Pomacentridae. The data found strong support for the monophyly of both the family Pomacentridae and the subfamily Amphiprioninae. Within the Amphiprioninae, Premnas was found nested within the genus Amphiprion, as the sister group to an Amphiprion ocellaris + A. percula clade, thereby rendering Amphiprion paraphyletic. The Chrominae were found to be polyphyletic, because one putative member (Mecaenichthys) appeared in a more basal position in the tree. Although the subfamily Chrominae was not found to be a monophyletic subfamily, there was strong support for the monophyly of a Chromis + Dascyllus clade. Within this clade, Chromis was found to be paraphyletic relative to Dascyllus. Tang s (2001) analysis also recovered a novel clade, one composed of the Amphiprioninae, Pomacentrus, Neoglyphidodon, and Amblyglyphidodon. This large clade was the sister group of the Chromis + Dascyllus clade. The Pomacentrinae were recovered as a broadly polyphyletic assemblage; putative members of this subfamily were scattered throughout the tree. There was strong support for the monophyly of four genera as well: Abudefduf, Dascyllus, Pomacentrus, and Stegastes. Abasal clade that included Stegastes, Microspathodon, Hypsypops, Parma, and Plectroglyphidodon was also recovered, although the support for this clade was weak. The goal of this study is to use sequence data from a portion of the 12S gene to infer the phylogenetic relationships among representative species of the family Pomacentridae, and to compare these results with the phylogenetic framework that has been proposed previously (Tang, 2001). One of the possible avenues of future research suggested by Tang (2001) was the addition of more taxa to improve sampling and representation, which this study accomplishes. This analysis will resolve the phylogenetic position of four genera and 25 species that were not represented in that previous study, providing a more robust estimate of pomacentrid phylogeny. 2. Materials and methods 2.1. Material examined Fishes used in this study were collected with a variety of methods, including the use of hand-nets while SCU- BAdiving, gillnets, and from aquarium specimens. Tissue samples were stored without preservation at )70 C in an ultracold freezer. Thirty-four species of pomacentrids were sequenced for this study, representing 14 of the 29 recognized genera. These taxa represent three of the four subfamilies; tissue from the monotypic subfamily Lepidozyginae was not available. Four species (Amphiprion frenatus, A. ocellaris, Dascyllus melanurus, and D. reticulatus) originally sequenced for this
3 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) project were first published in a previous analysis (Tang, 2001). Sequence data from an additional 19 taxa were included from Tang (2001). Thus, data from a total of 53 pomacentrids, representing 48 different species and 18 genera, were compiled for the data matrix used in this study. Five species (Abudefduf sexfasciatus, Amphiprion clarkii, Dascyllus aruanus, Plectroglyphidodon lacrymatus, and Premnas biaculeatus) were represented by two different individuals in the data matrix. Following Eschmeyer et al. (1998), the name Stegastes adustus (Troschel) is used herein as the senior synonym for S. dorsopunicans (Poey). Acomplete list of ingroup representatives examined is given in Table 1. Outgroup taxa were drawn from the Cichlidae, Embiotocidae, Labridae, and Moronidae. The first three families (Cichlidae, Embiotocidae, and Labridae) are outgroup candidates because they, along with the Pomacentridae, are considered members of the suborder Labroidei (Kaufman and Liem, 1982; Lauder and Liem, 1983; Stiassny and Jensen, 1987). However, considering the debate over the monophyly of the Labroidei (see Section 1), in addition to including a single species from each of those three putative labroid families as outgroups, one species from the family Moronidae was included as well to serve as a generic perciform outgroup; sequence data for this outgroup taxon were taken from Tang et al. (1999). Acomplete list of outgroup species examined is given in Table DNA amplification and sequencing Only a portion of the 12S gene region was examined for this study. This project, which only entailed the sequencing of the 12S region, was completed independently of the work done by Tang (2001), so data from the 16S gene were not collected. Despite the absence of 16S data, this data matrix has almost 60% more sequenced base positions (54,574 bp, prior to alignment) than what was sequenced in the previous study (34,121 bp, prior to alignment; Tang, 2001) because of the increased number of taxa. For the 34 taxa sequenced in this study, genomic extractions were done from muscle tissue with standard phenol/chloroform extraction protocols, following Kocher et al. (1989). Target regions of the mtdnawere amplified using the polymerase chain reaction (PCR), following Saiki (1990). TaqDNApolymerase (HT Biotechnology) and the two 20-bp oligonucleotide primers listed in Table 2 were used to amplify an approximately 600-bp fragment of the 12S ribosomal gene, using the following thermal cycling profile: 94 C denaturing for 60 s, 45 C annealing for 70 s, and 72 C extension for 2 min for 36 cycles (modified from Saiki, 1990). Each PCR reaction was preceded by a hot start at 94 C for 2 min to improve the yield. The resulting amplified products were then reamplified to generate single-stranded DNA(ssDNA), following Sanger et al. (1977). The resulting ssdna fragment was purified via agarose gel separation, electro-elution, and phenol/chloroform extraction, following Sambrook et al. (1989). The purified product was suspended in double-distilled water and prepared for manual sequencing, which was performed with an Amplicycle sequencing kit (Perkin Elmer), [ 35 S]-dNTPs, and the primers indicated in Table 2. The sequencing reactions were carried out with the following thermal cycling profile: 95 C for denaturing, 45 C for annealing, and 72 C for extension, 60 s for each step, for 30 cycles. Each sequencing reaction was electrophoresed on a 6% polyacrylamide gel and visualized by exposure to X-Omat X-ray film (Kodak) for h. The sequences ranged from 378 bp (Neopomacentrus azysron) to 646 bp (D. aruanus and P. biaculeatus) in length, prior to alignment. All sequences were deposited in GenBank (Table 1). All DNA sequence data from Tang (2001) were collected via automated sequencing. See Tang (2001) for the DNAamplification and sequencing protocols used for the other 19 pomacentrids and the four outgroup species that were used in this analysis DNA alignment All DNA sequences from the complementary light and heavy strands were spliced together to form a consensus light strand sequence. These 32 consensus sequences were first compiled into a data matrix using the Pileup option of the computer program GCG 8.01 (Genetic Computer Group, Wisconsin Sequence Analysis) and were subsequently aligned against a previously published data matrix (Tang, 2001). The published sequences were originally compiled using the computer program Sequence Navigator 1.01 (Applied Biosystems), where a preliminary alignment was performed using the CLUSTAL option included in that computer software. These data were then exported as a NEXUS file and the sequences were manually aligned in PAUP* 4.0b6 (Swofford, unpublished). The data were organized into stem and loop regions using published secondary structure models for the 12S mitochondrial ribosomal DNAgene (Van de Peer et al., 1994). See Tang (2001) for a more detailed treatment of sequence alignment procedures. All sequences generated for this study were incorporated into the existing data matrix used in that study. APAUP/NEXUS file with the complete alignment is available from the corresponding author upon request; the file may also be downloaded at Missing data There was incomplete overlap in the two sets of sequence data. The sequences produced for this analysis begin approximately 300 bp from the 5 0 end of the 12S
4 448 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) Table 1 List of species used in the study, following accepted classification (Allen, 1975, 1991), and GenBank accession numbers Taxon Catalog number (KU tissue number) Accession no. Order Perciformes Suborder Percoidel Family Moronidae Morone chrysops KU (T823) AF Suborder Labroidei Family Cichlidae Crenicichla lepidota KU (T605) AF Family Embiotocidae Embiotoca jacksoni SIO uncataloged (T58) AF Family Labridae Halichoeres chrysus KU (T29) AF Family Pomacentridae Subfamily Amphiprioninae Amphiprion clarkii KU (T49) AF A: clarkii #2 ASIZ uncataloged AF A: frenatus ASIZ uncataloged AF A: ocellaris ASIZ uncataloged AF A. percula KU (T2928) AF A: perideraion ASIZ uncataloged AF Premnas biaculeatus KU (T20) AF P: biaculeatus #2 ASIZ uncataloged AF Subfamily Chrominae Acanthochromis polyacanthus ASIZ uncataloged AF Chromis analis ASIZ uncataloged AF Chro. cyanea USNM (T77) AF Chro: fumea ASIZ uncataloged AF Chro. iomelas USNM (T704) AF Chro:viridis ASIZ uncataloged AF Dascyllus aruanus USNM (T768) AF D: aruanus #2 ASIZ uncataloged AF D: melanurus ASIZ uncataloged AF D: reticulatus ASIZ uncataloged AF Mecaenichthys immaculatus AMS (T3093) AF Subfamily Pomacentrinae Abudefduf saxatilis USNM (T194) AF A. sexfasciatus USNM (T714) AF A: sexfasciatus #2 ASIZ uncataloged AF A: sordidus ASIZ uncataloged AF A: vaigiensis ASIZ uncataloged AF Amblyglyphidodon aureus USNM (T773) AF A: curacao ASIZ uncataloged AF Chrysiptera leucopoma ASIZ uncataloged AF C: rex ASIZ uncataloged AF Hypsypops rubicundus KU (T3488) AF Microspathodon chrysurus USNM (T142) AF Neoglyphidodon melas ASIZ uncataloged AF N: nigroris ASIZ uncataloged AF N. polyacanthus AMS (T3094) AF Neopomacentrus azysron ASIZ uncataloged AF Parma oligolepis AMS (T3096) AF Plectroglyphidodon dickii ASIZ uncataloged AF P. lacrymatus USNM (T661) AF P: lacrymatus #2 ASIZ uncataloged AF P: leucozonus ASIZ uncataloged AF Pomacentrus auriventrus ASIZ uncataloged AF P: bankanensis ASIZ uncataloged AF P. brachialis USNM (T763) AF P: chrysurus ASIZ uncataloged AF P: coelestis ASIZ uncataloged AF P: moluccensis ASIZ uncataloged AF P. vaiuli USNM (T690) AF Stegastes adustus USNM (T86) AF S: altus ASIZ uncataloged AF081243
5 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) Table 1 (continued) Taxon Catalog number (KU tissue number) Accession No. S: fasciolatus ASIZ uncataloged AF S: lividus ASIZ uncataloged AF S: obreptus ASIZ uncataloged AF S. variabilis USNM (T188) AF Teixeirichthys jordani ASIZ uncataloged AF Note. Species in boldface text were sequenced for this study, all other materials examined are from Tang et al. (1999) and Tang (2001). Abbreviations. AMS, Australian Museum; ASIZ, Academia Sinica, Institute of Zoology, ROC; KU, University of Kansas; SIO, Scripps Institute of Oceanography; USNM, Smithsonian Institution, United States National Museum. Table 2 Sequencing and amplification primers used in the study Primer Sequence ( ) PB2 (L) CAAGTTGACAGACAACGGCG PV (H) GCACGGATGTCTTCTCGGTG gene and extend through to the 3 0 end of the 12S gene, whereas most of the sequences from Tang (2001) include the 5 0 end of the 12S gene, beginning approximately 50 bp from the 3 0 end of the trna-phe gene, but do not extend to the 3 0 end, stopping approximately 100 bp short of the start of the trna-val gene. Therefore, all the sequences from Tang (2001) were missing at least 100 bp from the 3 0 end of the 12S gene and all the new sequences generated are missing at least 300 bp from the 5 0 end of the gene. One species, Neopomacentrus azysron, is missing approximately 600 bp from the 5 0 end of the gene, and only 378 bp were collected from the 3 0 end because of an error during sequencing. Although all of the taxa have some characters with missing data, studies have shown that, given a choice, including such characters in an analysis is better than simply excluding all those characters because doing so generally increases phylogenetic accuracy, provided the missing data are not abundant (Wiens, 1998). Therefore, rather than deleting hundreds of base positions (aligned data columns) for which only some data were missing, those characters were coded as missing and included for the phylogenetic analysis Site saturation Analyses were performed to identify potential site saturation (i.e., multiple mutations at a single site) in both the stem and loop regions. Possible site saturation was examined by plotting the number of substitutions between pairs of taxa against mean patristic distance and Tamura Nei distance (Tamura and Nei, 1993); both the patristic distance and Tamura Nei values were generated in PAUP* 4.0 (Swofford, unpublished). Mean patristic distances were calculated from the 100 best trees retained in an initial heuristic parsimony search. The 100 patristic distances for each pairwise comparison were parsed and the mean was calculated by the computer program PatSat (Benson, unpublished) Phylogenetic analyses Phylogenetic analyses were carried out using the heuristic search option of PAUP* 4.0b6 (Swofford, unpublished), with 1000 random addition sequence replicates and tree-bisection-reconnection set as options. Gaps were common in loop regions and were included in all analyses as a fifth character state. All characters were equally weighted. Phylogenetic trees were evaluated using summary values reported by PAUP (e.g., tree length, consistency index). Support for each internode (i.e., monophyletic group) was evaluated by calculating decay index (Bremer, 1988, 1994) and bootstrap values (Felsenstein, 1985). Decay indices were generated using TreeRot (Sorenson, 1996) and bootstrap values were calculated in PAUP, using 1000 bootstrap replications of a simple heuristic search. Three constrained analyses were conducted to examine possible alternate topologies of relationships. The constraint trees enforced: (1) a monophyletic Chrominae; (2) a monophyletic Chromis; and (3) a monophyletic Stegastes. Templeton s tests (Templeton, 1983) were then performed to compare each of these alternate tree topologies to the most-parsimonious topologies. The character state differences were generated by MacClade 4.0 (Maddison and Maddison, 2000). The statistical tests were conducted with the computer program Minitab 10.5 (Minitab). 3. Results The final aligned data matrix contained 1058 aligned base positions. Five such data columns were excluded from the analyses because of ambiguous alignment (i.e., an inability to confidently make homology statements between different taxa). Of the remaining 1053 base positions, 383 were parsimony-informative. Plotting the number of differences between pairs of taxa against their Tamura Nei distance and mean patristic distance revealed no discernable evidence of site saturation in the stem regions or among the transversion substitutions in the loop regions. Among the least conservative class of mutations, transitions in loop regions, there was no conclusive evidence for site saturation. Plotting the number of loop transitions against Tamura Nei distance shows little or no deviation from a linear relation
6 450 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) Fig. 1. Scatter-plot of the number of transition substitutions vs. the (A) Tamura Nei distance and (B) mean patristic distance in pairwise comparisons in loop regions. between the number of mutations and sequence divergence (Fig. 1A). The mean patristic distance plot displays an ambiguous result, there may or may not be a divergence from a linear relationship (Fig. 1B). The phylogenetic analysis yielded 16 most-parsimonious trees each with a total length of 2124 steps (CI ¼ 0.435; HI ¼ 0.565; RI ¼ 0.643; RC ¼ 0.280). The strict consensus of the 16 trees is shown with decay index and bootstrap values at the appropriate internodes (Fig. 2). Bootstrap values below 50% are not shown. This strict consensus topology is well resolved, with only a few areas of uncertainty. The alternate topologies are entirely the result of instability within two clades, the genus Abudefduf and the subfamily Amphipioninae. The family itself, Pomacentridae, is recovered as a monophyletic group. Of the three subfamilies examined for this study, only the Amphiprioninae appears to be monophyletic. Within the Amphiprioninae, the position of Premnas is unstable relative to the species of Amphiprion: in four of the most-parsimonious trees Premnas is found to be the sister group to a monophyletic Amphiprion; in the other 12 trees, Premnas is recovered within the genus Amphiprion, as the sister group to an A. ocellaris + A. percula clade (subgenus Actinicola). Both the Chrominae and the Pomacentrinae are polyphyletic. Even though the subfamily Chrominae is not monophlyetic, there is support for a Chromis + Dascyllus clade (Fig. 2), but Chromis appears to be paraphyletic relative to a monophyletic Dascyllus. The other two chromine genera that were sequenced, Acanthochromis and Mecaenichthys (both monotypic genera), appear in very different parts of the tree. The subfamily Pomacentrinae is broadly polyphyletic, with putative members distributed throughout the tree. Among its genera, Abudefduf, Amblyglyphidodon, Chrysiptera, Neoglyphidodon, and Pomacentrus are monophyletic, with Abudefduf and Chrysiptera having the best branch support. Pomacentrus is recovered as the sister group to the Amphiprioninae and these taxa are part of a much larger clade which also includes Acanthochromis, Amblyglyphidodon, Chrysiptera, Neoglyphidodon, Neopomacentrus, and Teixeirichthys. The branch support for this large crown group is among the most robust in the tree, this group has the best support of any clade containing more than two genera. Within this group, Acanthochromis, a putative chromine, appears to be the sister group to an Amblyglyphidodon + Neoglyphidodon clade, and this clade of three genera is sister to the Amphiprioninae + Pomacentrus clade. Chrysiptera is the sister group of that clade, with Neopomacentrus as the next most basal member, and Teixeirichthys at the base of the large clade. The constrained analysis enforcing a monophyletic Stegastes resulted in trees that were one step longer (TL ¼ 2125) than the most-parsimonious resolution (TL ¼ 2124). The results of the Templeton s test indicated that this alternate topology was not significantly different from the most-parsimonious one ðp ¼ 0:40Þ. The constrained analysis enforcing a monophyletic Chrominae recovered trees that were 52 steps longer (TL ¼ 2176) than the shortest trees, and comparison of the shortest trees with the constrained trees indicated that the alternate topologies were significantly different ðp < 0:0001Þ. Trees 10 steps longer (TL ¼ 2134) resulted from the analysis enforcing a monophyletic Chromis, and the Templeton s test found that this topology was significantly different ðp ¼ 0:02Þ.
7 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) Fig. 2. Strict consensus of 16 most-parsimonious trees, total length ¼ 2124 steps, CI ¼ 0.435, HI ¼ 0.565, RI ¼ 0.643, RC ¼ (values are for each most-parsimonious tree). Decay index support (above) and bootstrap values for 1000 replicates (below) are indicated at each node, bootstrap values below 50% have been omitted. 4. Discussion Sixteen most-parsimonious trees resulted from the phylogenetic analysis. The strict consensus of those most-parsimonious topologies finds a monophyletic family Pomacentridae (Fig. 2). Monophyly of the family is congruent with previous hypotheses, both molecular (Tang, 2001) and morphological (Fitzpatrick, 1992; Kaufman and Liem, 1982; Stiassny, 1981). Support for a monophyletic Pomacentridae was strong in the previous molecular analysis (Tang, 2001), and this tree topology is highly congruent with the relationships proposed in that study. Of the three subfamilies examined herein, the subfamily Amphiprioninae (anemonefishes) is the only one that was found to be monophyletic. Support for this clade is relatively weak, which is surprising considering previous studies have found strong support for the
8 452 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) monophyly of this group: phylogenetic analyses using morphological data find that the group is united by a number of synapomorphies (Fitzpatrick, 1992; Tang, unpublished), and molecular studies have corroborated those findings (Elliott et al., 1999; Tang, 2001). Though they did not include many outgroup pomacentrids in their analysis, Elliott et al. (1999) found support for a monophyletic Amphiprioninae. Tang (2001) included additional genera and species of pomacentrids, and found very robust support for the monophyly of the subfamily. Within the Amphiprioninae, the relationships are largely unresolved. In 12 of the 16 most-parsimonious trees, Amphiprion is not recovered as monophyletic because Premnas appears as the sister group to the subgenus Actinicola (A. ocellaris + A. percula). However, in four of the most-parsimonious resolutions, Amphiprion is monophyletic, with Premnas as its sister genus. These two conflicting results correspond with the two current hypotheses of anemonefish relationships. Amonophyletic Amphiprion agrees with the preferred topology of Elliott et al. (1999, Fig. 3a), whereas a paraphyletic Amphiprion, with Premnas as the sister to an A. ocellaris + A. percula clade (subgenus Actinicola) agrees with the tree recovered by Tang (2001, Fig. 2). Such a sistergroup relationship between the two species of Actinicola (the percula species complex of Allen, 1972) and Premnas has been proposed previously by Allen (1972, Fig. 12). Even though Elliott et al. s (1999) preferred tree shows a monophyletic Amphiprion, some of their cytochrome b data supports an A. ocellaris + Premnas clade (Elliott et al., 1999, pp ). The data resolving this part of the tree are not conclusive, additional data are required to settle this issue. These results resolve a monophyletic subgenus Actinicola (A. ocellaris + A. percula) within Amphiprion, with strong branch support, an outcome congruent with the classification proposed by Allen (1972) and the results of Tang (2001). Other relationships within the genus Amphiprion do not match those of previous studies. For example, the monophyly of the subgenus Amphiprion is not supported, which agrees with the results of Elliott et al. (1999); however, relationships among the species are not the same as those results. There is strong support for a clade composed of A. clarkii, A. frenatus, and A. perideraion, and two representatives of A. clarkii group together as well. Without more representatives, conclusions about Amphiprion relationships are tentative at best. There is strong support for the monophyly of a large crown clade composed of the Amphiprioninae, Pomacentrus, Acanthochromis, Amblyglyphidodon, Neoglyphidodon, Chrysiptera, Neopomacentrus, andteixeirichthys (Fig. 2). The relationships recovered in this clade suggest several novel relationships which are unlike anything that has been proposed in traditional classifications of the family (Allen, 1972, 1975). However, there is corroboration for such a clade from the previous molecular phylogeny of this group (Tang, 2001). That study did not have all the taxa examined for this analysis; however, all those that were included (Amphiprion, Premnas, Pomacentrus, Amblyglyphidodon, and Neoglyphidodon) did form a clade, with strong branch support (Tang, 2001, Fig. 2). The sister group of the subfamily Amphiprioninae appears to be Pomacentrus, a monophyletic genus. This relationship corroborates the results of Tang (2001), which found an Amphiprioninae + Pomacentrus clade. The subfamily Chrominae is polyphyletic and there is good evidence for this. From the Templeton s test, a tree enforcing the monophyly of the subfamily based on these data would be not only much longer (52 steps) than the shortest tree, but also significantly different from that shortest tree ðp < 0:0001Þ. However, despite the polyphyly of the Chrominae, a substantial subset of the group does cluster together. There is strong support for a Chromis + Dascyllus clade, with some of the more robustly supported branches in the tree found within the genus Dascyllus. The topology of relationships found within Dascyllus is identical to the one found in Tang (2001), which is not surprising since that study included the same four species examined here; the only new addition is a second representative of D. aruanus. The nominate genus of the subfamily, Chromis, is paraphyletic relative to Dascyllus. This finding appears to be non-trivial as the branch uniting C. iomelas with the Dascyllus clade is well supported. The results of the Templeton s test provide additional evidence supporting this, as constraining a monophyletic Chromis would yield a tree significantly different from the most-parsimonious resolution. Aparaphyletic Chromis is consistent with the results of Tang (2001), which had two Chromis representatives and found a similar result. Larval characteristics lend some support to this as well. Although larval characters have not been used before in a phylogenetic analysis, Chromis species display a great diversity among its larvae, a diversity of larval forms not seen in other pomacentrid groups (Kavanagh et al., 2000). The other large clade found by this analysis is a basal one containing an assortment of different genera: Hypsypops, Microspathodon, Parma, Plectroglyphidodon (in part), and Stegastes. The presence of such a clade is corroborated by Tang (2001), which found a clade with a similar composition of genera, though with fewer representative species and a monophyletic Stegastes. The genus Stegastes appears polyphyletic in this analysis, but support in this part of the tree is weak; trees only one step away from the most-parsimonious trees recover a monophyletic Stegastes, making it hard to draw any conclusions about the status of Stegastes with confidence. Not surprisingly, the Templeton s test highlights
9 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) this problem, as constraining a monophyletic Stegastes results in trees that are not significantly different from the most-parsimonious ones. In general, support in this basal part of the tree is not robust. Aside from the two representatives of Plectroglyphidodon lacrymatus grouping together and a Stegastes adustus + S. variabilis clade, no branches have very strong support. Overall, the relationships recovered in this analysis corroborate many of the findings of Tang (2001), though the two studies differ in several details. Both trees recover monophyletic Pomacentridae and Amphiprioninae, as well as monophyletic Abudefduf, Dascyllus, and Pomacentrus. Both studies find a clade which contains the Amphiprioninae, Amblyglyphidodon, Neoglyphidodon, and Pomacentrus, with Pomacentrus as the sister group of the Amphiprioninae. Both studies also agree on a sister-group relationship between this large clade and a Chromis + Dascyllus clade, and both find Chromis paraphyletic relative to Dascyllus Taxonomic implications The results of this study support several relationships that do not agree with the traditional classification of the family Pomacentridae, though the trees yielded from this study largely corroborate the earlier results of Tang (2001). The most obvious difference between the relationships based on molecular data and the traditional hypothesis involves the status of the subfamilies within the Pomacentridae. Though only three of the four putative subfamilies were included in this study, it is clear that some of them do not accurately reflect monophyletic groups within the family. The one subfamily not examined, Lepidozyginae, is monotypic. Of the remaining three, only one, the Amphiprioninae, appears to be monophyletic. The other two are polyphyletic, with putative species of both subfamilies scattered in different parts of the tree. This situation presents some difficult taxonomic problems. The Chrominae likely will require a reduction in its constituent species. Currently, only the species of Chromis and Dascyllus form a monophyletic group; even the monophyly of Chromis itself is in doubt and the relationships of the other two chromine genera, Altrichthys and Azurina, which were not available for sequencing, are unknown. The far flung nature of the pomacentrine genera in the tree poses a larger problem. The nature of the tree topology does not appear to allow any easy division of the pomacentrine genera into convenient monophyletic subfamilies. Chief among these problems is that the nominate genus for the subfamily, Pomacentrus, is the sister group of the only monophyletic subfamily, the Amphiprioninae. Although this is far from a complete sampling of pomacentrid taxa, it seems unlikely that the addition of new taxa and data will drastically change the topology, rendering broadly polyphyletic groups like the Pomacentrinae monophyletic. Given this, eventually, when a more complete hypothesis of relationships within the family is available, a complete revision of this family and its component subfamilies will be necessary. More genera and species need to be examined, not only to establish the relationships within the family and subfamilies, but also to test the monophyly of speciose genera like Amphiprion, Chromis, Chrysiptera, Pomacentrus, and Stegastes. Acknowledgments We thank K.-M. Chen, C.-H. Ho, and L.-T. Ho for their help in collecting the specimens used in the study. C.-G. Sang, W.-B. Yeh, and J.-P. Chen provided invaluable technical assistance. We thank M.J. Grose, N.I. Holcroft, C.A. Sheil, E.O. Wiley, and two anonymous reviewers for their suggestions and comments. Collection of some sequence data in this study was partly funded by grant DEB from the National Science Foundation (USA). References Allen, G.R., Anemonefishes. T.F.H. Publications, Neptune City, NJ. Allen, G.R., Damselfishes of the South Seas. T.F.H. Publications, Neptune City, NJ. Allen, G.R., Freshwater Fishes of Australia. T.F.H. Publications, Neptune City, NJ. Allen, G.R., Damselfishes of the World. Aquarium Systems, Melle, Germany. Bremer, K., The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42, Bremer, K., Branch support and tree stability. Cladistics 10, Elliott, J.K., Lougheed, S., Bateman, B., McPhee, L., Boag, P., Molecular phylogenetic evidence for the evolution of specialization in anemonefishes. Proc. R. Soc. London B 266, Eschmeyer, W.N., Ferraris Jr., C.J., Hoang, M.D., Long, D.J., Species of fishes. In: Eschmeyer, W.N. (Ed.), Catalog of Fishes. California Academy of Sciences, San Francisco, pp Felsenstein, J., Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, Fitzpatrick, S., Pomacentrid intrafamilial relationships: a cladistic approach. Unpublished Honours thesis, James Cook University, Townsville, Queensland, Australia. Johnson, G.D., Percomorph phylogeny: progress and problems. Bull. Mar. Sci. 52, Kaufman, L.S., Liem, K.F., Fishes of the suborder Labroidei (Pisces: Perciformes): phylogeny, ecology and evolutionary significance. Breviora 472, Kavanagh, K.D., Leis, J.M., Rennis, D.S., Pomacentridae. In: Leis, J.M., Carson-Ewart, B.M. (Eds.), The larvae of Indo-Pacific coastal fishes: an identification guide to marine fish larvae. Brill Academic Publishers, Boston, pp Kocher, T.D., Thomas, W.K., Meyer, A., Edwards, S.V., P a abo, S., Villablanca, F.X., Wilson, A.C., Dynamics of mitochondrial DNAevolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 86,
10 454 N.-H. Jang-Liawet al. / Molecular Phylogenetics and Evolution 25 (2002) Lauder, G.V., Liem, K.F., The evolution and interrelationships of the actinopterygian fishes. Bull. Mus. Comp. Zool. 150, Maddison, D.R., Maddison, W.P., MacClade 4.0. Sinauer Associates, Sunderland, MA. Nelson, J.S., Fishes of the World, third ed. John Wiley and Sons, New York. Norman, J.R., ADraft Synopsis of the Orders, Families and Genera of Recent Fishes and Fish-like Vertebrates. Trustees of the British Museum, London. Rosen, D.E., Patterson, C., On M uller s and Cuvier s concepts of pharyngognath and labyrinth fishes and the classification of percomorph fishes, with an atlas of percomorph dorsal gill arches. Am. Mus. Nov. 2983, Saiki, R.K., Amplification of genomic DNA. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J. (Eds.), PCR Protocols. A Guide to Methods and Applications. Academic Press, New York, pp Sambrook, J., Fritsch, E.F., Maniatis, T., Molecular Cloning: A Laboratory Manual, second ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. Sanger, F., Nicklen, S., Coulson, A.R., DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, Shao, K.T., Numerical taxonomic study on damselfishes (Pisces: Pomacentridae) II. Generic interrelationships from morphometric studies. In: Proceedings of the Symposium on Marine Biological Science, Biology Research Center. National Science Council Monograph Series 14, pp Sorenson, M.D., TreeRot. University of Michigan, Ann Arbor, MI. Stiassny, M.L.J., The phyletic status of the family Cichlidae (Pisces: Perciformes): a comparative anatomical investigation. Neth. J. Zool. 31, Stiassny, M.L.J., Jensen, J.S., Labroid interrelationships revisited: morphological complexity, key innovations, and the study of comparative diversity. Bull. Mus. Comp. Zool. 151, Streelman, J.T., Karl, S.A., Reconstructing labroid evolution with single-copy nuclear DNA. Proc. R. Soc. Lond. B 264, Swofford, D.L., unpublished. PAUP* 4.0b6. Tamura, T., Nei, M., Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNAin humans and chimpanzees. Mol. Biol. Evol. 10, Tang, K.L., Phylogenetic relationships among damselfishes (Teleostei: Pomacentridae) as determined by mitochondrial DNA data. Copeia 2001 (3), Tang, K.L., Berendzen, P.B., Wiley, E.O., Morrissey, J.F., Winterbottom, R., Johnson, G.D., The phylogenetic relationships of the suborder Acanthuroidei (Teleostei: Perciformes) based on molecular and morphological evidence. Mol. Phyl. Evol. 11, Templeton, A.R., Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of apes and humans. Evolution 37, Van de Peer, Y., Van den Broeck, I., De Rijk, P., De Wachter, R., Database on the structure of small subunit RNA. Nucleic Acids Res. 22, Wiens, J.J., Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst. Biol. 47,
Lecture 11 Wednesday, September 19, 2012
 Lecture 11 Wednesday, September 19, 2012 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean
Lecture 11 Wednesday, September 19, 2012 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean
Species: Panthera pardus Genus: Panthera Family: Felidae Order: Carnivora Class: Mammalia Phylum: Chordata
 CHAPTER 6: PHYLOGENY AND THE TREE OF LIFE AP Biology 3 PHYLOGENY AND SYSTEMATICS Phylogeny - evolutionary history of a species or group of related species Systematics - analytical approach to understanding
CHAPTER 6: PHYLOGENY AND THE TREE OF LIFE AP Biology 3 PHYLOGENY AND SYSTEMATICS Phylogeny - evolutionary history of a species or group of related species Systematics - analytical approach to understanding
Fig Phylogeny & Systematics
 Fig. 26- Phylogeny & Systematics Tree of Life phylogenetic relationship for 3 clades (http://evolution.berkeley.edu Fig. 26-2 Phylogenetic tree Figure 26.3 Taxonomy Taxon Carolus Linnaeus Species: Panthera
Fig. 26- Phylogeny & Systematics Tree of Life phylogenetic relationship for 3 clades (http://evolution.berkeley.edu Fig. 26-2 Phylogenetic tree Figure 26.3 Taxonomy Taxon Carolus Linnaeus Species: Panthera
CLADISTICS Student Packet SUMMARY Phylogeny Phylogenetic trees/cladograms
 CLADISTICS Student Packet SUMMARY PHYLOGENETIC TREES AND CLADOGRAMS ARE MODELS OF EVOLUTIONARY HISTORY THAT CAN BE TESTED Phylogeny is the history of descent of organisms from their common ancestor. Phylogenetic
CLADISTICS Student Packet SUMMARY PHYLOGENETIC TREES AND CLADOGRAMS ARE MODELS OF EVOLUTIONARY HISTORY THAT CAN BE TESTED Phylogeny is the history of descent of organisms from their common ancestor. Phylogenetic
Bio 1B Lecture Outline (please print and bring along) Fall, 2006
 Bio 1B Lecture Outline (please print and bring along) Fall, 2006 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #4 -- Phylogenetic Analysis (Cladistics) -- Oct.
Bio 1B Lecture Outline (please print and bring along) Fall, 2006 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #4 -- Phylogenetic Analysis (Cladistics) -- Oct.
Rostral Horn Evolution Among Agamid Lizards of the Genus. Ceratophora Endemic to Sri Lanka
 Rostral Horn Evolution Among Agamid Lizards of the Genus Ceratophora Endemic to Sri Lanka James A. Schulte II 1, J. Robert Macey 2, Rohan Pethiyagoda 3, Allan Larson 1 1 Department of Biology, Box 1137,
Rostral Horn Evolution Among Agamid Lizards of the Genus Ceratophora Endemic to Sri Lanka James A. Schulte II 1, J. Robert Macey 2, Rohan Pethiyagoda 3, Allan Larson 1 1 Department of Biology, Box 1137,
HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS
 HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS WASHINGTON AND LONDON 995 by the Smithsonian Institution All rights reserved
HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS WASHINGTON AND LONDON 995 by the Smithsonian Institution All rights reserved
Clownfish Guide READ ONLINE
 Clownfish Guide READ ONLINE The Gold Nugget Maroon Clownfish is ORA's most distinct clownfish to date. Its unique appearance manifests itself similarly to the Platinum Percula. The clown fish (also known
Clownfish Guide READ ONLINE The Gold Nugget Maroon Clownfish is ORA's most distinct clownfish to date. Its unique appearance manifests itself similarly to the Platinum Percula. The clown fish (also known
muscles (enhancing biting strength). Possible states: none, one, or two.
 Reconstructing Evolutionary Relationships S-1 Practice Exercise: Phylogeny of Terrestrial Vertebrates In this example we will construct a phylogenetic hypothesis of the relationships between seven taxa
Reconstructing Evolutionary Relationships S-1 Practice Exercise: Phylogeny of Terrestrial Vertebrates In this example we will construct a phylogenetic hypothesis of the relationships between seven taxa
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution
 Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
Modern Evolutionary Classification. Lesson Overview. Lesson Overview Modern Evolutionary Classification
 Lesson Overview 18.2 Modern Evolutionary Classification THINK ABOUT IT Darwin s ideas about a tree of life suggested a new way to classify organisms not just based on similarities and differences, but
Lesson Overview 18.2 Modern Evolutionary Classification THINK ABOUT IT Darwin s ideas about a tree of life suggested a new way to classify organisms not just based on similarities and differences, but
Testing Phylogenetic Hypotheses with Molecular Data 1
 Testing Phylogenetic Hypotheses with Molecular Data 1 How does an evolutionary biologist quantify the timing and pathways for diversification (speciation)? If we observe diversification today, the processes
Testing Phylogenetic Hypotheses with Molecular Data 1 How does an evolutionary biologist quantify the timing and pathways for diversification (speciation)? If we observe diversification today, the processes
Phylogeny Reconstruction
 Phylogeny Reconstruction Trees, Methods and Characters Reading: Gregory, 2008. Understanding Evolutionary Trees (Polly, 2006) Lab tomorrow Meet in Geology GY522 Bring computers if you have them (they will
Phylogeny Reconstruction Trees, Methods and Characters Reading: Gregory, 2008. Understanding Evolutionary Trees (Polly, 2006) Lab tomorrow Meet in Geology GY522 Bring computers if you have them (they will
Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1
 Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1 Systematics is the comparative study of biological diversity with the intent of determining the relationships between organisms. Humankind has always
Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1 Systematics is the comparative study of biological diversity with the intent of determining the relationships between organisms. Humankind has always
History of Lineages. Chapter 11. Jamie Oaks 1. April 11, Kincaid Hall 524. c 2007 Boris Kulikov boris-kulikov.blogspot.
 History of Lineages Chapter 11 Jamie Oaks 1 1 Kincaid Hall 524 joaks1@gmail.com April 11, 2014 c 2007 Boris Kulikov boris-kulikov.blogspot.com History of Lineages J. Oaks, University of Washington 1/46
History of Lineages Chapter 11 Jamie Oaks 1 1 Kincaid Hall 524 joaks1@gmail.com April 11, 2014 c 2007 Boris Kulikov boris-kulikov.blogspot.com History of Lineages J. Oaks, University of Washington 1/46
These small issues are easily addressed by small changes in wording, and should in no way delay publication of this first- rate paper.
 Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper reports on a highly significant discovery and associated analysis that are likely to be of broad interest to the scientific community.
Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper reports on a highly significant discovery and associated analysis that are likely to be of broad interest to the scientific community.
Horned lizard (Phrynosoma) phylogeny inferred from mitochondrial genes and morphological characters: understanding conflicts using multiple approaches
 Molecular Phylogenetics and Evolution xxx (2004) xxx xxx MOLECULAR PHYLOGENETICS AND EVOLUTION www.elsevier.com/locate/ympev Horned lizard (Phrynosoma) phylogeny inferred from mitochondrial genes and morphological
Molecular Phylogenetics and Evolution xxx (2004) xxx xxx MOLECULAR PHYLOGENETICS AND EVOLUTION www.elsevier.com/locate/ympev Horned lizard (Phrynosoma) phylogeny inferred from mitochondrial genes and morphological
17.2 Classification Based on Evolutionary Relationships Organization of all that speciation!
 Organization of all that speciation! Patterns of evolution.. Taxonomy gets an over haul! Using more than morphology! 3 domains, 6 kingdoms KEY CONCEPT Modern classification is based on evolutionary relationships.
Organization of all that speciation! Patterns of evolution.. Taxonomy gets an over haul! Using more than morphology! 3 domains, 6 kingdoms KEY CONCEPT Modern classification is based on evolutionary relationships.
Introduction to Cladistic Analysis
 3.0 Copyright 2008 by Department of Integrative Biology, University of California-Berkeley Introduction to Cladistic Analysis tunicate lamprey Cladoselache trout lungfish frog four jaws swimbladder or
3.0 Copyright 2008 by Department of Integrative Biology, University of California-Berkeley Introduction to Cladistic Analysis tunicate lamprey Cladoselache trout lungfish frog four jaws swimbladder or
LABORATORY EXERCISE 6: CLADISTICS I
 Biology 4415/5415 Evolution LABORATORY EXERCISE 6: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Biology 4415/5415 Evolution LABORATORY EXERCISE 6: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
The phylogeny of antiarch placoderms. Sarah Kearsley Geology 394 Senior Thesis
 The phylogeny of antiarch placoderms Sarah Kearsley Geology 394 Senior Thesis Abstract The most comprehensive phylogenetic study of antiarchs to date (Zhu, 1996) included information not derived from observation.
The phylogeny of antiarch placoderms Sarah Kearsley Geology 394 Senior Thesis Abstract The most comprehensive phylogenetic study of antiarchs to date (Zhu, 1996) included information not derived from observation.
PARTIAL REPORT. Juvenile hybrid turtles along the Brazilian coast RIO GRANDE FEDERAL UNIVERSITY
 RIO GRANDE FEDERAL UNIVERSITY OCEANOGRAPHY INSTITUTE MARINE MOLECULAR ECOLOGY LABORATORY PARTIAL REPORT Juvenile hybrid turtles along the Brazilian coast PROJECT LEADER: MAIRA PROIETTI PROFESSOR, OCEANOGRAPHY
RIO GRANDE FEDERAL UNIVERSITY OCEANOGRAPHY INSTITUTE MARINE MOLECULAR ECOLOGY LABORATORY PARTIAL REPORT Juvenile hybrid turtles along the Brazilian coast PROJECT LEADER: MAIRA PROIETTI PROFESSOR, OCEANOGRAPHY
GEODIS 2.0 DOCUMENTATION
 GEODIS.0 DOCUMENTATION 1999-000 David Posada and Alan Templeton Contact: David Posada, Department of Zoology, 574 WIDB, Provo, UT 8460-555, USA Fax: (801) 78 74 e-mail: dp47@email.byu.edu 1. INTRODUCTION
GEODIS.0 DOCUMENTATION 1999-000 David Posada and Alan Templeton Contact: David Posada, Department of Zoology, 574 WIDB, Provo, UT 8460-555, USA Fax: (801) 78 74 e-mail: dp47@email.byu.edu 1. INTRODUCTION
1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters
 1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. The sister group of J. K b. The sister group
1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. The sister group of J. K b. The sister group
LABORATORY EXERCISE 7: CLADISTICS I
 Biology 4415/5415 Evolution LABORATORY EXERCISE 7: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Biology 4415/5415 Evolution LABORATORY EXERCISE 7: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
UNIT III A. Descent with Modification(Ch19) B. Phylogeny (Ch20) C. Evolution of Populations (Ch21) D. Origin of Species or Speciation (Ch22)
 UNIT III A. Descent with Modification(Ch9) B. Phylogeny (Ch2) C. Evolution of Populations (Ch2) D. Origin of Species or Speciation (Ch22) Classification in broad term simply means putting things in classes
UNIT III A. Descent with Modification(Ch9) B. Phylogeny (Ch2) C. Evolution of Populations (Ch2) D. Origin of Species or Speciation (Ch22) Classification in broad term simply means putting things in classes
A Mitochondrial DNA Phylogeny of Extant Species of the Genus Trachemys with Resulting Taxonomic Implications
 NOTES AND FIELD REPORTS 131 Chelonian Conservation and Biology, 2008, 7(1): 131 135 Ó 2008 Chelonian Research Foundation A Mitochondrial DNA Phylogeny of Extant Species of the Genus Trachemys with Resulting
NOTES AND FIELD REPORTS 131 Chelonian Conservation and Biology, 2008, 7(1): 131 135 Ó 2008 Chelonian Research Foundation A Mitochondrial DNA Phylogeny of Extant Species of the Genus Trachemys with Resulting
Rostral Horn Evolution among Agamid Lizards of the Genus Ceratophora Endemic to Sri Lanka
 Molecular Phylogenetics and Evolution Vol. 22, No. 1, January, pp. 111 117, 2002 doi:10.1006/mpev.2001.1041, available online at http://www.idealibrary.com on Rostral Horn Evolution among Agamid Lizards
Molecular Phylogenetics and Evolution Vol. 22, No. 1, January, pp. 111 117, 2002 doi:10.1006/mpev.2001.1041, available online at http://www.idealibrary.com on Rostral Horn Evolution among Agamid Lizards
Title: Phylogenetic Methods and Vertebrate Phylogeny
 Title: Phylogenetic Methods and Vertebrate Phylogeny Central Question: How can evolutionary relationships be determined objectively? Sub-questions: 1. What affect does the selection of the outgroup have
Title: Phylogenetic Methods and Vertebrate Phylogeny Central Question: How can evolutionary relationships be determined objectively? Sub-questions: 1. What affect does the selection of the outgroup have
1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters
 1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. Identify the taxon (or taxa if there is more
1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. Identify the taxon (or taxa if there is more
The Making of the Fittest: LESSON STUDENT MATERIALS USING DNA TO EXPLORE LIZARD PHYLOGENY
 The Making of the Fittest: Natural The The Making Origin Selection of the of Species and Fittest: Adaptation Natural Lizards Selection in an Evolutionary and Adaptation Tree INTRODUCTION USING DNA TO EXPLORE
The Making of the Fittest: Natural The The Making Origin Selection of the of Species and Fittest: Adaptation Natural Lizards Selection in an Evolutionary and Adaptation Tree INTRODUCTION USING DNA TO EXPLORE
Evolution of Biodiversity
 Long term patterns Evolution of Biodiversity Chapter 7 Changes in biodiversity caused by originations and extinctions of taxa over geologic time Analyses of diversity in the fossil record requires procedures
Long term patterns Evolution of Biodiversity Chapter 7 Changes in biodiversity caused by originations and extinctions of taxa over geologic time Analyses of diversity in the fossil record requires procedures
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes)
 Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes) Phylogenetics is the study of the relationships of organisms to each other.
Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes) Phylogenetics is the study of the relationships of organisms to each other.
The melanocortin 1 receptor (mc1r) is a gene that has been implicated in the wide
 Introduction The melanocortin 1 receptor (mc1r) is a gene that has been implicated in the wide variety of colors that exist in nature. It is responsible for hair and skin color in humans and the various
Introduction The melanocortin 1 receptor (mc1r) is a gene that has been implicated in the wide variety of colors that exist in nature. It is responsible for hair and skin color in humans and the various
What are taxonomy, classification, and systematics?
 Topic 2: Comparative Method o Taxonomy, classification, systematics o Importance of phylogenies o A closer look at systematics o Some key concepts o Parts of a cladogram o Groups and characters o Homology
Topic 2: Comparative Method o Taxonomy, classification, systematics o Importance of phylogenies o A closer look at systematics o Some key concepts o Parts of a cladogram o Groups and characters o Homology
INQUIRY & INVESTIGATION
 INQUIRY & INVESTIGTION Phylogenies & Tree-Thinking D VID. UM SUSN OFFNER character a trait or feature that varies among a set of taxa (e.g., hair color) character-state a variant of a character that occurs
INQUIRY & INVESTIGTION Phylogenies & Tree-Thinking D VID. UM SUSN OFFNER character a trait or feature that varies among a set of taxa (e.g., hair color) character-state a variant of a character that occurs
Name: Date: Hour: Fill out the following character matrix. Mark an X if an organism has the trait.
 Name: Date: Hour: CLADOGRAM ANALYSIS What is a cladogram? It is a diagram that depicts evolutionary relationships among groups. It is based on PHYLOGENY, which is the study of evolutionary relationships.
Name: Date: Hour: CLADOGRAM ANALYSIS What is a cladogram? It is a diagram that depicts evolutionary relationships among groups. It is based on PHYLOGENY, which is the study of evolutionary relationships.
Phylogeny of genus Vipio latrielle (Hymenoptera: Braconidae) and the placement of Moneilemae group of Vipio species based on character weighting
 International Journal of Biosciences IJB ISSN: 2220-6655 (Print) 2222-5234 (Online) http://www.innspub.net Vol. 3, No. 3, p. 115-120, 2013 RESEARCH PAPER OPEN ACCESS Phylogeny of genus Vipio latrielle
International Journal of Biosciences IJB ISSN: 2220-6655 (Print) 2222-5234 (Online) http://www.innspub.net Vol. 3, No. 3, p. 115-120, 2013 RESEARCH PAPER OPEN ACCESS Phylogeny of genus Vipio latrielle
Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per.
 Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per. Introduction Imagine a single diagram representing the evolutionary relationships between everything that has ever lived. If life evolved
Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per. Introduction Imagine a single diagram representing the evolutionary relationships between everything that has ever lived. If life evolved
Cladistics (reading and making of cladograms)
 Cladistics (reading and making of cladograms) Definitions Systematics The branch of biological sciences concerned with classifying organisms Taxon (pl: taxa) Any unit of biological diversity (eg. Animalia,
Cladistics (reading and making of cladograms) Definitions Systematics The branch of biological sciences concerned with classifying organisms Taxon (pl: taxa) Any unit of biological diversity (eg. Animalia,
DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS
 Syst. Biol. 45(4):39^14, 1996 DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS STEVEN POE Department of Zoology and Texas Memorial Museum, University of Texas, Austin, Texas 78712-1064, USA; E-mail:
Syst. Biol. 45(4):39^14, 1996 DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS STEVEN POE Department of Zoology and Texas Memorial Museum, University of Texas, Austin, Texas 78712-1064, USA; E-mail:
Phylogeographic assessment of Acanthodactylus boskianus (Reptilia: Lacertidae) based on phylogenetic analysis of mitochondrial DNA.
 Zoology Department Phylogeographic assessment of Acanthodactylus boskianus (Reptilia: Lacertidae) based on phylogenetic analysis of mitochondrial DNA By HAGAR IBRAHIM HOSNI BAYOUMI A thesis submitted in
Zoology Department Phylogeographic assessment of Acanthodactylus boskianus (Reptilia: Lacertidae) based on phylogenetic analysis of mitochondrial DNA By HAGAR IBRAHIM HOSNI BAYOUMI A thesis submitted in
Systematics, Taxonomy and Conservation. Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem
 Systematics, Taxonomy and Conservation Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem What is expected of you? Part I: develop and print the cladogram there
Systematics, Taxonomy and Conservation Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem What is expected of you? Part I: develop and print the cladogram there
The impact of the recognizing evolution on systematics
 The impact of the recognizing evolution on systematics 1. Genealogical relationships between species could serve as the basis for taxonomy 2. Two sources of similarity: (a) similarity from descent (b)
The impact of the recognizing evolution on systematics 1. Genealogical relationships between species could serve as the basis for taxonomy 2. Two sources of similarity: (a) similarity from descent (b)
AP Lab Three: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST
 AP Biology Name AP Lab Three: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST In the 1990 s when scientists began to compile a list of genes and DNA sequences in the human genome
AP Biology Name AP Lab Three: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST In the 1990 s when scientists began to compile a list of genes and DNA sequences in the human genome
Spacing pattern and body size composition of the protandrous anemonefish Amphiprion frenatus inhabiting colonial host anemones
 Spacing pattern and body size composition of the protandrous anemonefish Amphiprion frenatus inhabiting colonial host anemones Miyako Kobayashi 1 and Akihisa Hattori 2* 1 Nature Conservation Educators
Spacing pattern and body size composition of the protandrous anemonefish Amphiprion frenatus inhabiting colonial host anemones Miyako Kobayashi 1 and Akihisa Hattori 2* 1 Nature Conservation Educators
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST In this laboratory investigation, you will use BLAST to compare several genes, and then use the information to construct a cladogram.
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST In this laboratory investigation, you will use BLAST to compare several genes, and then use the information to construct a cladogram.
6. The lifetime Darwinian fitness of one organism is greater than that of another organism if: A. it lives longer than the other B. it is able to outc
 1. The money in the kingdom of Florin consists of bills with the value written on the front, and pictures of members of the royal family on the back. To test the hypothesis that all of the Florinese $5
1. The money in the kingdom of Florin consists of bills with the value written on the front, and pictures of members of the royal family on the back. To test the hypothesis that all of the Florinese $5
Animal Diversity III: Mollusca and Deuterostomes
 Animal Diversity III: Mollusca and Deuterostomes Objectives: Be able to identify specimens from the main groups of Mollusca and Echinodermata. Be able to distinguish between the bilateral symmetry on a
Animal Diversity III: Mollusca and Deuterostomes Objectives: Be able to identify specimens from the main groups of Mollusca and Echinodermata. Be able to distinguish between the bilateral symmetry on a
Ch 1.2 Determining How Species Are Related.notebook February 06, 2018
 Name 3 "Big Ideas" from our last notebook lecture: * * * 1 WDYR? Of the following organisms, which is the closest relative of the "Snowy Owl" (Bubo scandiacus)? a) barn owl (Tyto alba) b) saw whet owl
Name 3 "Big Ideas" from our last notebook lecture: * * * 1 WDYR? Of the following organisms, which is the closest relative of the "Snowy Owl" (Bubo scandiacus)? a) barn owl (Tyto alba) b) saw whet owl
Global comparisons of beta diversity among mammals, birds, reptiles, and amphibians across spatial scales and taxonomic ranks
 Journal of Systematics and Evolution 47 (5): 509 514 (2009) doi: 10.1111/j.1759-6831.2009.00043.x Global comparisons of beta diversity among mammals, birds, reptiles, and amphibians across spatial scales
Journal of Systematics and Evolution 47 (5): 509 514 (2009) doi: 10.1111/j.1759-6831.2009.00043.x Global comparisons of beta diversity among mammals, birds, reptiles, and amphibians across spatial scales
TOPIC CLADISTICS
 TOPIC 5.4 - CLADISTICS 5.4 A Clades & Cladograms https://upload.wikimedia.org/wikipedia/commons/thumb/4/46/clade-grade_ii.svg IB BIO 5.4 3 U1: A clade is a group of organisms that have evolved from a common
TOPIC 5.4 - CLADISTICS 5.4 A Clades & Cladograms https://upload.wikimedia.org/wikipedia/commons/thumb/4/46/clade-grade_ii.svg IB BIO 5.4 3 U1: A clade is a group of organisms that have evolved from a common
Evolution of Agamidae. species spanning Asia, Africa, and Australia. Archeological specimens and other data
 Evolution of Agamidae Jeff Blackburn Biology 303 Term Paper 11-14-2003 Agamidae is a family of squamates, including 53 genera and over 300 extant species spanning Asia, Africa, and Australia. Archeological
Evolution of Agamidae Jeff Blackburn Biology 303 Term Paper 11-14-2003 Agamidae is a family of squamates, including 53 genera and over 300 extant species spanning Asia, Africa, and Australia. Archeological
Systematics and taxonomy of the genus Culicoides what is coming next?
 Systematics and taxonomy of the genus Culicoides what is coming next? Claire Garros 1, Bruno Mathieu 2, Thomas Balenghien 1, Jean-Claude Delécolle 2 1 CIRAD, Montpellier, France 2 IPPTS, Strasbourg, France
Systematics and taxonomy of the genus Culicoides what is coming next? Claire Garros 1, Bruno Mathieu 2, Thomas Balenghien 1, Jean-Claude Delécolle 2 1 CIRAD, Montpellier, France 2 IPPTS, Strasbourg, France
Caecilians (Gymnophiona)
 Caecilians (Gymnophiona) David J. Gower* and Mark Wilkinson Department of Zoology, The Natural History Museum, London SW7 5BD, UK *To whom correspondence should be addressed (d.gower@nhm. ac.uk) Abstract
Caecilians (Gymnophiona) David J. Gower* and Mark Wilkinson Department of Zoology, The Natural History Museum, London SW7 5BD, UK *To whom correspondence should be addressed (d.gower@nhm. ac.uk) Abstract
NATIONAL BIORESOURCE DEVELOPMENT BOARD Dept. of Biotechnology Government of India, New Delhi
 NATIONAL BIORESOURCE DEVELOPMENT BOARD Dept. of Biotechnology Government of India, New Delhi MARINE BIORESOURCES FORMS DATA ENTRY: Form- 1(general ) (please answer only relevant fields;add additional fields
NATIONAL BIORESOURCE DEVELOPMENT BOARD Dept. of Biotechnology Government of India, New Delhi MARINE BIORESOURCES FORMS DATA ENTRY: Form- 1(general ) (please answer only relevant fields;add additional fields
Warm-Up: Fill in the Blank
 Warm-Up: Fill in the Blank 1. For natural selection to happen, there must be variation in the population. 2. The preserved remains of organisms, called provides evidence for evolution. 3. By using and
Warm-Up: Fill in the Blank 1. For natural selection to happen, there must be variation in the population. 2. The preserved remains of organisms, called provides evidence for evolution. 3. By using and
Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST
 Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST INVESTIGATION 3 BIG IDEA 1 Lab Investigation 3: BLAST Pre-Lab Essential Question: How can bioinformatics be used as a tool to
Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST INVESTIGATION 3 BIG IDEA 1 Lab Investigation 3: BLAST Pre-Lab Essential Question: How can bioinformatics be used as a tool to
Postilla PEABODY MUSEUM OF NATURAL HISTORY YALE UNIVERSITY NEW HAVEN, CONNECTICUT, U.S.A.
 Postilla PEABODY MUSEUM OF NATURAL HISTORY YALE UNIVERSITY NEW HAVEN, CONNECTICUT, U.S.A. Number 117 18 March 1968 A 7DIAPSID (REPTILIA) PARIETAL FROM THE LOWER PERMIAN OF OKLAHOMA ROBERT L. CARROLL REDPATH
Postilla PEABODY MUSEUM OF NATURAL HISTORY YALE UNIVERSITY NEW HAVEN, CONNECTICUT, U.S.A. Number 117 18 March 1968 A 7DIAPSID (REPTILIA) PARIETAL FROM THE LOWER PERMIAN OF OKLAHOMA ROBERT L. CARROLL REDPATH
PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, NY 10024
 PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, NY 10024 Number 3365, 61 pp., 7 figures, 3 tables May 17, 2002 Phylogenetic Relationships of Whiptail Lizards
PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, NY 10024 Number 3365, 61 pp., 7 figures, 3 tables May 17, 2002 Phylogenetic Relationships of Whiptail Lizards
Lineage Classification of Canine Title Disorders Using Mitochondrial DNA 宮原, 和郎, 鈴木, 三義. Journal of Veterinary Medical Sci Citation
 ' ' Lineage Classification of Canine Title Disorders Using Mitochondrial DNA TAKAHASI, Shoko, MIYAHARA, Kazuro Author(s) Hirosi, ISHIGURO, Naotaka, SUZUKI 宮原, 和郎, 鈴木, 三義 Journal of Veterinary Medical Sci
' ' Lineage Classification of Canine Title Disorders Using Mitochondrial DNA TAKAHASI, Shoko, MIYAHARA, Kazuro Author(s) Hirosi, ISHIGURO, Naotaka, SUZUKI 宮原, 和郎, 鈴木, 三義 Journal of Veterinary Medical Sci
Comparing DNA Sequences Cladogram Practice
 Name Period Assignment # See lecture questions 75, 122-123, 127, 137 Comparing DNA Sequences Cladogram Practice BACKGROUND Between 1990 2003, scientists working on an international research project known
Name Period Assignment # See lecture questions 75, 122-123, 127, 137 Comparing DNA Sequences Cladogram Practice BACKGROUND Between 1990 2003, scientists working on an international research project known
No limbs Eastern glass lizard. Monitor lizard. Iguanas. ANCESTRAL LIZARD (with limbs) Snakes. No limbs. Geckos Pearson Education, Inc.
 No limbs Eastern glass lizard Monitor lizard guanas ANCESTRAL LZARD (with limbs) No limbs Snakes Geckos Species: Panthera pardus Genus: Panthera Family: Felidae Order: Carnivora Class: Mammalia Phylum:
No limbs Eastern glass lizard Monitor lizard guanas ANCESTRAL LZARD (with limbs) No limbs Snakes Geckos Species: Panthera pardus Genus: Panthera Family: Felidae Order: Carnivora Class: Mammalia Phylum:
HONR219D Due 3/29/16 Homework VI
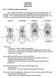 Part 1: Yet More Vertebrate Anatomy!!! HONR219D Due 3/29/16 Homework VI Part 1 builds on homework V by examining the skull in even greater detail. We start with the some of the important bones (thankfully
Part 1: Yet More Vertebrate Anatomy!!! HONR219D Due 3/29/16 Homework VI Part 1 builds on homework V by examining the skull in even greater detail. We start with the some of the important bones (thankfully
Comparing DNA Sequence to Understand
 Comparing DNA Sequence to Understand Evolutionary Relationships with BLAST Name: Big Idea 1: Evolution Pre-Reading In order to understand the purposes and learning objectives of this investigation, you
Comparing DNA Sequence to Understand Evolutionary Relationships with BLAST Name: Big Idea 1: Evolution Pre-Reading In order to understand the purposes and learning objectives of this investigation, you
Subdomain Entry Vocabulary Modules Evaluation
 Subdomain Entry Vocabulary Modules Evaluation Technical Report Vivien Petras August 11, 2000 Abstract: Subdomain entry vocabulary modules represent a way to provide a more specialized retrieval vocabulary
Subdomain Entry Vocabulary Modules Evaluation Technical Report Vivien Petras August 11, 2000 Abstract: Subdomain entry vocabulary modules represent a way to provide a more specialized retrieval vocabulary
Molecular Phylogenetics and Evolution
 Molecular Phylogenetics and Evolution 59 (2011) 623 635 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev A multigenic perspective
Molecular Phylogenetics and Evolution 59 (2011) 623 635 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev A multigenic perspective
NAUSHONIA PAN AMEN SIS, NEW SPECIES (DECAPODA: THALASSINIDEA: LAOMEDIIDAE) FROM THE PACIFIC COAST OF PANAMA, WITH NOTES ON THE GENUS
 5 October 1982 PROC. BIOL. SOC. WASH. 95(3), 1982, pp. 478-483 NAUSHONIA PAN AMEN SIS, NEW SPECIES (DECAPODA: THALASSINIDEA: LAOMEDIIDAE) FROM THE PACIFIC COAST OF PANAMA, WITH NOTES ON THE GENUS Joel
5 October 1982 PROC. BIOL. SOC. WASH. 95(3), 1982, pp. 478-483 NAUSHONIA PAN AMEN SIS, NEW SPECIES (DECAPODA: THALASSINIDEA: LAOMEDIIDAE) FROM THE PACIFIC COAST OF PANAMA, WITH NOTES ON THE GENUS Joel
PERSPECTIVES IN MARICULTURE
 PERSPECTIVES IN MARICULTURE Editors N. G. Menon and P. P. Pillai Central Marine Fisheries Research Institute, Cochin The Imveitigator The Marine Biological Association of India Post Box No. 1604, Tatapuram
PERSPECTIVES IN MARICULTURE Editors N. G. Menon and P. P. Pillai Central Marine Fisheries Research Institute, Cochin The Imveitigator The Marine Biological Association of India Post Box No. 1604, Tatapuram
Genetic diversity of the Indo-Pacific barrel sponge Xestospongia testudinaria (Haplosclerida : Petrosiidae)
 9 th World Sponge Conference 2013. 4-8 November 2013, Fremantle WA, Australia Genetic diversity of the Indo-Pacific barrel sponge Xestospongia testudinaria (Haplosclerida : Petrosiidae) Edwin Setiawan
9 th World Sponge Conference 2013. 4-8 November 2013, Fremantle WA, Australia Genetic diversity of the Indo-Pacific barrel sponge Xestospongia testudinaria (Haplosclerida : Petrosiidae) Edwin Setiawan
DO BROWN-HEADED COWBIRDS LAY THEIR EGGS AT RANDOM IN THE NESTS OF RED-WINGED BLACKBIRDS?
 Wilson Bull., 0(4), 989, pp. 599605 DO BROWNHEADED COWBIRDS LAY THEIR EGGS AT RANDOM IN THE NESTS OF REDWINGED BLACKBIRDS? GORDON H. ORTANS, EIVIN RDSKAPT, AND LES D. BELETSKY AssrnAcr.We tested the hypothesis
Wilson Bull., 0(4), 989, pp. 599605 DO BROWNHEADED COWBIRDS LAY THEIR EGGS AT RANDOM IN THE NESTS OF REDWINGED BLACKBIRDS? GORDON H. ORTANS, EIVIN RDSKAPT, AND LES D. BELETSKY AssrnAcr.We tested the hypothesis
Selection, Recombination and History in a Parasitic Flatworm (Echinococcus) Inferred from Nucleotide Sequences
 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 93(5): 695-702, Sep./Oct. 1998 Selection, Recombination and History in a Parasitic Flatworm (Echinococcus) Inferred from Nucleotide Sequences KL Haag, AM Araújo,
Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 93(5): 695-702, Sep./Oct. 1998 Selection, Recombination and History in a Parasitic Flatworm (Echinococcus) Inferred from Nucleotide Sequences KL Haag, AM Araújo,
Understanding Evolutionary History: An Introduction to Tree Thinking
 1 Understanding Evolutionary History: An Introduction to Tree Thinking Laura R. Novick Kefyn M. Catley Emily G. Schreiber Vanderbilt University Western Carolina University Vanderbilt University Version
1 Understanding Evolutionary History: An Introduction to Tree Thinking Laura R. Novick Kefyn M. Catley Emily G. Schreiber Vanderbilt University Western Carolina University Vanderbilt University Version
Molecular study for the sex identification in Japanese quails (Coturnix Japonica) Iran.
 Molecular study for the sex identification in Japanese quails (Coturnix Japonica) Nasrollah Vali1 1 and Abbas Doosti 2 1 Department of Animal Sciences, Faculty of Agriculture, Islamic Azad University,
Molecular study for the sex identification in Japanese quails (Coturnix Japonica) Nasrollah Vali1 1 and Abbas Doosti 2 1 Department of Animal Sciences, Faculty of Agriculture, Islamic Azad University,
Living Planet Report 2018
 Living Planet Report 2018 Technical Supplement: Living Planet Index Prepared by the Zoological Society of London Contents The Living Planet Index at a glance... 2 What is the Living Planet Index?... 2
Living Planet Report 2018 Technical Supplement: Living Planet Index Prepared by the Zoological Society of London Contents The Living Planet Index at a glance... 2 What is the Living Planet Index?... 2
Analysis of CR1 repeats in the zebra finch genome
 Analysis of CR1 repeats in the zebra finch genome George E. Liu, Yali Hou* and Twain Brown Bovine Functional Genomics Laboratory, ANRI, ARS, USDA, Beltsville, Maryland 20705, USA *Also affiliated with
Analysis of CR1 repeats in the zebra finch genome George E. Liu, Yali Hou* and Twain Brown Bovine Functional Genomics Laboratory, ANRI, ARS, USDA, Beltsville, Maryland 20705, USA *Also affiliated with
Evolution in Action: Graphing and Statistics
 Evolution in Action: Graphing and Statistics OVERVIEW This activity serves as a supplement to the film The Origin of Species: The Beak of the Finch and provides students with the opportunity to develop
Evolution in Action: Graphing and Statistics OVERVIEW This activity serves as a supplement to the film The Origin of Species: The Beak of the Finch and provides students with the opportunity to develop
Clarifications to the genetic differentiation of German Shepherds
 Clarifications to the genetic differentiation of German Shepherds Our short research report on the genetic differentiation of different breeding lines in German Shepherds has stimulated a lot interest
Clarifications to the genetic differentiation of German Shepherds Our short research report on the genetic differentiation of different breeding lines in German Shepherds has stimulated a lot interest
Sparse Supermatrices for Phylogenetic Inference: Taxonomy, Alignment, Rogue Taxa, and the Phylogeny of Living Turtles
 Syst. Biol. 59(1):42 58, 2010 c The Author(s) 2009. Published by Oxford University Press, on behalf of the Society of Systematic Biologists. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Syst. Biol. 59(1):42 58, 2010 c The Author(s) 2009. Published by Oxford University Press, on behalf of the Society of Systematic Biologists. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Are node-based and stem-based clades equivalent? Insights from graph theory
 Are node-based and stem-based clades equivalent? Insights from graph theory November 18, 2010 Tree of Life 1 2 Jeremy Martin, David Blackburn, E. O. Wiley 1 Associate Professor of Mathematics, San Francisco,
Are node-based and stem-based clades equivalent? Insights from graph theory November 18, 2010 Tree of Life 1 2 Jeremy Martin, David Blackburn, E. O. Wiley 1 Associate Professor of Mathematics, San Francisco,
2008/048 Reducing Dolphin Bycatch in the Pilbara Finfish Trawl Fishery
 2008/048 Reducing Dolphin Bycatch in the Pilbara Finfish Trawl Fishery PRINCIPAL INVESTIGATOR: Prof. N.R. Loneragan ADDRESS: Centre for Fish and Fisheries Research Biological Sciences and Biotechnology
2008/048 Reducing Dolphin Bycatch in the Pilbara Finfish Trawl Fishery PRINCIPAL INVESTIGATOR: Prof. N.R. Loneragan ADDRESS: Centre for Fish and Fisheries Research Biological Sciences and Biotechnology
May 10, SWBAT analyze and evaluate the scientific evidence provided by the fossil record.
 May 10, 2017 Aims: SWBAT analyze and evaluate the scientific evidence provided by the fossil record. Agenda 1. Do Now 2. Class Notes 3. Guided Practice 4. Independent Practice 5. Practicing our AIMS: E.3-Examining
May 10, 2017 Aims: SWBAT analyze and evaluate the scientific evidence provided by the fossil record. Agenda 1. Do Now 2. Class Notes 3. Guided Practice 4. Independent Practice 5. Practicing our AIMS: E.3-Examining
SUPPLEMENTARY INFORMATION
 In comparison to Proganochelys (Gaffney, 1990), Odontochelys semitestacea is a small turtle. The adult status of the specimen is documented not only by the generally well-ossified appendicular skeleton
In comparison to Proganochelys (Gaffney, 1990), Odontochelys semitestacea is a small turtle. The adult status of the specimen is documented not only by the generally well-ossified appendicular skeleton
Molecular Systematics and Evolution of Regina and the Thamnophiine Snakes
 Molecular Phylogenetics and Evolution Vol. 21, No. 3, December, pp. 408 423, 2001 doi:10.1006/mpev.2001.1024, available online at http://www.idealibrary.com on Molecular Systematics and Evolution of Regina
Molecular Phylogenetics and Evolution Vol. 21, No. 3, December, pp. 408 423, 2001 doi:10.1006/mpev.2001.1024, available online at http://www.idealibrary.com on Molecular Systematics and Evolution of Regina
Do the traits of organisms provide evidence for evolution?
 PhyloStrat Tutorial Do the traits of organisms provide evidence for evolution? Consider two hypotheses about where Earth s organisms came from. The first hypothesis is from John Ray, an influential British
PhyloStrat Tutorial Do the traits of organisms provide evidence for evolution? Consider two hypotheses about where Earth s organisms came from. The first hypothesis is from John Ray, an influential British
A phylogeny for side-necked turtles (Chelonia: Pleurodira) based on mitochondrial and nuclear gene sequence variation
 Bivlogkal Journal ofthe Linnean So&& (1998), 67: 2 13-246. \\'ith 4 figures Article ID biji.1998.0300, avaiiable online at http://www.idealihrary.lom on IDE kt @ c A phylogeny for side-necked turtles (Chelonia:
Bivlogkal Journal ofthe Linnean So&& (1998), 67: 2 13-246. \\'ith 4 figures Article ID biji.1998.0300, avaiiable online at http://www.idealihrary.lom on IDE kt @ c A phylogeny for side-necked turtles (Chelonia:
Evolutionary Trade-Offs in Mammalian Sensory Perceptions: Visual Pathways of Bats. By Adam Proctor Mentor: Dr. Emma Teeling
 Evolutionary Trade-Offs in Mammalian Sensory Perceptions: Visual Pathways of Bats By Adam Proctor Mentor: Dr. Emma Teeling Visual Pathways of Bats Purpose Background on mammalian vision Tradeoffs and bats
Evolutionary Trade-Offs in Mammalian Sensory Perceptions: Visual Pathways of Bats By Adam Proctor Mentor: Dr. Emma Teeling Visual Pathways of Bats Purpose Background on mammalian vision Tradeoffs and bats
Required and Recommended Supporting Information for IUCN Red List Assessments
 Required and Recommended Supporting Information for IUCN Red List Assessments This is Annex 1 of the Rules of Procedure for IUCN Red List Assessments 2017 2020 as approved by the IUCN SSC Steering Committee
Required and Recommended Supporting Information for IUCN Red List Assessments This is Annex 1 of the Rules of Procedure for IUCN Red List Assessments 2017 2020 as approved by the IUCN SSC Steering Committee
Diagnosis of Living and Fossil Short-necked Turtles of the Genus Elseya using skeletal morphology
 Diagnosis of Living and Fossil Short-necked Turtles of the Genus Elseya using skeletal morphology by Scott Andrew Thomson B.App.Sc. University of Canberra Institute of Applied Ecology University of Canberra
Diagnosis of Living and Fossil Short-necked Turtles of the Genus Elseya using skeletal morphology by Scott Andrew Thomson B.App.Sc. University of Canberra Institute of Applied Ecology University of Canberra
Biology 120 Lab Exam 2 Review
 Biology 120 Lab Exam 2 Review Student Learning Services and Biology 120 Peer Mentors Sunday, November 26 th, 2017 4:00 pm Arts 263 Important note: This review was written by your Biology Peer Mentors (not
Biology 120 Lab Exam 2 Review Student Learning Services and Biology 120 Peer Mentors Sunday, November 26 th, 2017 4:00 pm Arts 263 Important note: This review was written by your Biology Peer Mentors (not
Turtles (Testudines) Abstract
 Turtles (Testudines) H. Bradley Shaffer Department of Evolution and Ecology, University of California, Davis, CA 95616, USA (hbshaffer@ucdavis.edu) Abstract Living turtles and tortoises consist of two
Turtles (Testudines) H. Bradley Shaffer Department of Evolution and Ecology, University of California, Davis, CA 95616, USA (hbshaffer@ucdavis.edu) Abstract Living turtles and tortoises consist of two
Morphological systematics of kingsnakes, Lampropeltis getula complex (Serpentes: Colubridae), in the eastern United States
 Zootaxa : 1 39 (2006) www.mapress.com/zootaxa/ Copyright 2006 Magnolia Press ISSN 1175-5326 (print edition) ZOOTAXA ISSN 1175-5334 (online edition) Morphological systematics of kingsnakes, Lampropeltis
Zootaxa : 1 39 (2006) www.mapress.com/zootaxa/ Copyright 2006 Magnolia Press ISSN 1175-5326 (print edition) ZOOTAXA ISSN 1175-5334 (online edition) Morphological systematics of kingsnakes, Lampropeltis
The Rufford Foundation Final Report
 The Rufford Foundation Final Report Congratulations on the completion of your project that was supported by The Rufford Foundation. We ask all grant recipients to complete a Final Report Form that helps
The Rufford Foundation Final Report Congratulations on the completion of your project that was supported by The Rufford Foundation. We ask all grant recipients to complete a Final Report Form that helps
Points of View Tetrapod Phylogeny, Amphibian Origins, and the De nition of the Name Tetrapoda
 Points of View Syst. Biol. 51(2):364 369, 2002 Tetrapod Phylogeny, Amphibian Origins, and the De nition of the Name Tetrapoda MICHEL LAURIN Équipe Formations squelettiques UMR CNRS 8570, Case 7077, Université
Points of View Syst. Biol. 51(2):364 369, 2002 Tetrapod Phylogeny, Amphibian Origins, and the De nition of the Name Tetrapoda MICHEL LAURIN Équipe Formations squelettiques UMR CNRS 8570, Case 7077, Université
HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION
 Syst. Zool., 3(3), 98, pp. 229-249 HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION DANIEL R. BROOKS Abstract Brooks, ID. R. (Department of Zoology, University of British Columbia, 275 Wesbrook Mall,
Syst. Zool., 3(3), 98, pp. 229-249 HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION DANIEL R. BROOKS Abstract Brooks, ID. R. (Department of Zoology, University of British Columbia, 275 Wesbrook Mall,
AKC Canine Health Foundation Grant Updates: Research Currently Being Sponsored By The Vizsla Club of America Welfare Foundation
 AKC Canine Health Foundation Grant Updates: Research Currently Being Sponsored By The Vizsla Club of America Welfare Foundation GRANT PROGRESS REPORT REVIEW Grant: 00748: SNP Association Mapping for Canine
AKC Canine Health Foundation Grant Updates: Research Currently Being Sponsored By The Vizsla Club of America Welfare Foundation GRANT PROGRESS REPORT REVIEW Grant: 00748: SNP Association Mapping for Canine
Phylogeny of Harpacticoida (Copepoda): Revision of Maxillipedasphalea and Exanechentera
 Phylogeny of Harpacticoida (Copepoda): Revision of Maxillipedasphalea and Exanechentera Sybille Seifried sybille.seifried@mail.uni-oldenburg.de published 2003 by Cuvillier Verlag, Göttingen ISBN 3-89873-845-0
Phylogeny of Harpacticoida (Copepoda): Revision of Maxillipedasphalea and Exanechentera Sybille Seifried sybille.seifried@mail.uni-oldenburg.de published 2003 by Cuvillier Verlag, Göttingen ISBN 3-89873-845-0
click for previous page SEA TURTLES
 click for previous page SEA TURTLES FAO Sheets Fishing Area 51 TECHNICAL TERMS AND PRINCIPAL MEASUREMENTS USED head width (Straight-line distances) head prefrontal precentral carapace central (or neural)
click for previous page SEA TURTLES FAO Sheets Fishing Area 51 TECHNICAL TERMS AND PRINCIPAL MEASUREMENTS USED head width (Straight-line distances) head prefrontal precentral carapace central (or neural)
Phylogeny of the Sciaroidea (Diptera): the implication of additional taxa and character data
 Zootaxa : 63 68 (2006) www.mapress.com/zootaxa/ Copyright 2006 Magnolia Press ISSN 1175-5326 (print edition) ZOOTAXA ISSN 1175-5334 (online edition) Phylogeny of the Sciaroidea (Diptera): the implication
Zootaxa : 63 68 (2006) www.mapress.com/zootaxa/ Copyright 2006 Magnolia Press ISSN 1175-5326 (print edition) ZOOTAXA ISSN 1175-5334 (online edition) Phylogeny of the Sciaroidea (Diptera): the implication
