HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION
|
|
|
- Homer Armstrong
- 5 years ago
- Views:
Transcription
1 Syst. Zool., 3(3), 98, pp HENNIG'S PARASITOLOGICAL METHOD: A PROPOSED SOLUTION DANIEL R. BROOKS Abstract Brooks, ID. R. (Department of Zoology, University of British Columbia, 275 Wesbrook Mall, Vancouver, B.C., Canada V6T 2A9) 98. Hennig's parasitological method: a proposed solution. Syst. Zool., 3: A quantitative solution for Hennig's parasitological method is presented. Cladograms summarizing natural relationships among parasite taxa are converted into host-group characters by means of additive binary coding. Unrooted Wagner analysis followed by most parsimonious rooting produces a maximum information-content representation of natural host-parasite relationships. Because host-parasite relationships result either from random colonization or from co-speciation, host relationships well corroborated by a multi-parasite analysis correspond to host phylogeny. Poorly corroborated host relationships indicate an ambiguous parasite message alerting a worker to possible host transfers. Thus, such analyses point out co-speciation and random colonization components of host-parasite systems. Single or multiple parasite taxa may be used. A host phylogeny based on non-parasite characters is neither necessary nor sufficient for studying phylogenetic aspects of coevolution, although such may be helpful in testing ambiguous aspects. Once a host-group cladogram based on parasites has been established, phylogenetic interpretations for each observed host-parasite relationship may be made according to a listed set of necessary and sufficient criteria. Finally, evaluation of two models of coevolution, a "vicariance" model and the "resource-tracking" model, indicates that the latter cannot be extrapolated successfully to explain congruent phylogenetic differentiation of hosts and parasites and that the former model represents the general pattern of natural relationships among hosts and parasites. [Cladistics; coevolution; phylogeny; quantitative systematics.] The non-random association between hosts and parasites has led many evolutionary biologists to conclude that parasites should in some way serve as markers of evolutionary relationships among hosts. Hennig (966:6-3, 74-8) discussed the problem under the rubric of "the parasitological method" and much of his analysis concerned the pitfalls of using parasite data when host and parasite groups had not experienced concomitant phylogenesis throughout their evolutionary histories. Because of the problems he outlined, Hennig suggested that parasite data constituted adjunct or subsidiary information to be applied a posteriori in confirming the correctness of any phylogenetic analysis. "[the parasitological method] has many points of contact with the chorological method..." (p. 3) and, "Particularly favorable results can be expected if the parasitological method is supported by geographic vicariance relationships." (p. 2) 229 Hennig's conclusions about the utility of parasitological data were not optimistic, "... the theory of the parasitological method must be much better worked out than it is now... it is a question of the degree of phylogenetic relationships, of deciding whether the host species are more closely related to one another than to species in which the parasites in question do not occur. Consequently, as with the morphological method, there must be criteria for determining when this assumption is justified... and when it is not. No such criteria are known." (p. 2) However, despite "... the present unsatisfactory state of its theoretical basis..." (p. 2), Hennig viewed the parasitological method as having great potential. "We cannot forsee the role that the parasitological method will play some day when relationships are more accurately known... We can be sure that it will not be small." (p. 8) Hennig was not a parasitologist so it is not surprising that he viewed the problem only from the host-group's viewpoint. He never wrote about the possi- Downloaded from at Penn State University (Paterno Lib) on May 2, 26
2 23 SYSTEMATIC ZOOLOGY VOL. 3 bility of formulating parasite cladograms separately, or of the possibility of comparing both host and parasite cladograms. I have been interested in the phylogenetic relationships of parasites, including their host and geographic relationships through time. I discovered that it was possible to construct cladograms of parasite relationships without using data on host relationships or geographical distributions. Thus, parasites have synapomorphies independent of their hosts just as hosts have synapomorphies independent of their parasites. Comparisons of host and parasite cladograms have been used as aids in reconstructing evolutionary histories of parasite taxa (Brooks, 977, 978a, 978b, 979b, 98; Brooks, Mayes, and Thorson, 98; Brooks and Overstreet, 978; Deardorff, Brooks, and Thorson, 98). Such studies can also be viewed as initial estimates of the kinds of characters various parasite taxa would provide for analyses of host phylogenies. They are necessary components of the proposed solution to Hennig's parasitological method presented herein, in which parasites will be considered characters of hosts. I will not be comparing the congruence of two or more phylogenetic trees. My considerations of the solution to the parasitological method and its implications for studies in coevolution comprise three stages: ) analytical protocol, including (a) one parasite taxon compared with the host-group cladogram, (b) all (or many) parasite taxa in a fauna or community compared with the hostgroup cladogram, and (c) all (or many) parasite taxa analyzed when there is no host-group cladogram available for comparisons; 2) phylogenetic interpretations of host-parasite relationships postulated by results of the analytical technique; and 3) evaluation of evolutionary scenarios for co-evolution. ANALYTICAL PROTOCOL What is the significance of finding a parasite in a particular host? That parasite may occur in the host as a result of a long-standing phylogenetic relationship between the host-group and the parasite group co-speciation (Brooks, 979a). Alternatively, the parasite may occur in a given host because it has colonized that host from another through some form of ecological homoplasy between the hosts relevant to the infective stage of the parasite. In the first case, depending on whether or not the hosts' phylogenesis and the parasites' phylogenesis occurred together, the parasite's occurrence might relate to the current host(s) or to the current host(s)'s ancestor; in the first instance, the parasite's occurrence is an apomorphic trait and in the second it is a plesiomorphic trait. Parasites thus exhibit plesiomorphous, apomorphous, and homoplasious relationships with their hosts and can be used as characters in the same manner as any other kind of characters. Parasitologists have been aware, at least implicitly, of this state of affairs. Manter (966) stated, "Parasites... furnish information about present-day habits and ecology of their individual hosts. These same parasites also hold promise of telling us something about host and geographical connections of long ago. They are simultaneously the product of an immediate environment and of a long ancestry reflecting associations of millions of years. The messages they carry are thus always bilingual and usually garbled." In phylogenetic analysis of any kind, garbled messages result from failure to distinguish homoplasious from homologous traits and generalized from specialized homologous traits. Phylogeneticists suggest that phylogenetic analyses involve evaluation of the states and polarities of each character used, utilization of as many characters as possible in formulating a phylogenetic hypothesis, and testing of the hypothesis by comparison with new sets of data. Manter's (966) prescription for evaluating the phylogenetic relationships of host-parasite systems was the following: "() each species of parasite should be evaluated separately; (2) as large a variety of parasites as Downloaded from at Penn State University (Paterno Lib) on May 2, 26
3 98 HENNIG S PARASITOLOGICAL METHOD FlGS. -6. Phylogenetic interpretation of the distribution among members of a host taxon of a single parasite taxon. Host-group is represented by the cladogram and parasite group by + and O symbols (presence and absence, respectively). Figure demonstrates symplesiomorphic occurrence of the parasite; Figure 2 demonstrates synapomorphic occurrence of the parasite; Figure 3 demonstrates convergent loss of the parasite; Figure 4 demonstrates convergent acquisition of the parasite; Figure 5 demonstrates apomorphic acquisition and secondary loss of the parasite (a form of reversal); and Figure 6 demonstrates apomorphic loss and secondary acquisition of the parasite (also a fonn of reversal). possible should be considered to offset errors or cases of convergence; and (3) conclusions should take into consideration other lines of evidence." If the parasite taxon is used as a single unit the presence and absence of that taxon in various hosts can be indicated directly on the host cladogram and interpretation of its distribution among hosts would follow the protocol shown in Figures -6, following the parsimony criterion of Farris (974; see also Platnick, 977) but require a host cladogram designated as true. However, if a number of terminal taxa in a parasite taxon are being used in comparisons, one cannot represent adequately the information content of the parasite cladogram (the unit of comparison) by superimposing parasite terminal taxa on the cladogram of host relationships. Nor can that information be represented by listing presence/absence data for the parasites and clustering hosts on the basis of shared presences and absences (a form of raw similarity clustering). I will demonstrate the problems with the above approach using two examples. Consider the following parasite cladogram (Fig. 7) and host cladogram (Fig. 8). It is clear by inspection that the two branching diagrams are identical this is a case in which there has been strict cospeciation (Brooks, 979:32, example IA). Figure 9 depicts a matrix of presence/absence data for the parasite terminal taxa A-D which occur in hosts I-IV, respectively. The resultant clustering diagram (Fig. ) does not represent adequately the coevolutionary relationships of the host group and parasite group. Raw similarity clustering of presence/absence data would provide the correct diagram only under a special set of data. Consider now a different set of parasites and of hosts (Figs., 2, respectively). Only if hosts I and II were inhabited by parasites A, B, and C, host III by parasites A and B, and host IV by parasite A would presence/absence data (Fig. 3) provide the right information (Fig. 4). There is still another means of han- Downloaded from at Penn State University (Paterno Lib) on May 2, 26
4 232 SYSTEMATIC ZOOLOGY VOL. 3 A B III IV III IV 5 A B C D II B III IV III IV I II III I V IV FiGS Using presence/absence data in studies of coevolution. Figures 7 and represent parasite taxa with terminal taxa denoted by letters. Figures 8 and 2 represent host taxa with terminal taxa denoted by Roman numerals. Matrices in Figures 9 and 3 depict presence/absence data for the various parasite terminal taxa as they occur in various hosts. Cladograms based on those matrices are shown in Figures and 4. Presence/absence data for Figure gives incomplete resolution of host relationships; such data in Figure 4 give complete resolution. In Figures 7-, parasite A occurs in host I, parasite B in host II, parasite C in host III, and parasite D in host IV. In Figures -4, parasite C occurs in hosts I and II, parasite B in hosts I, II, and III, and parasite A in hosts I, II, III, and IV. dling parasite data, one which preserves all the information under any set of hostparasite relationships. That method involves treating the parasite cladogram as if it were a character-state tree, labeling A B C D E F G 9 A B C D E 2 7 FiGS Using phylogenetic relationships of parasites in revolutionary studies. Figures 5 and 9 correspond to Figures 7 and but have internal nodes labeled. Figures 6 and 2 correspond to Figures 8 and 2. Figures 7 and 2 represent matrices of parasite data based on Additive Binary Coding of parasite cladograms. Rows correspond to terminal taxa and nodes from original cladogram; columns correspond to new binary characters. Figures 8 and 22 depict host cladograms based on matrices in Figures 7 and 2. Host-parasite relationships are the same as indicated in Figures 7- and -4. the internal nodes, and converting all terminal taxa and nodes into a matrix of binary characters which represent the cladogram exactly. This is a use of the technique of Additive Binary Coding used by phylogeneticists to analyze character-state trees or to standardize multistate characters with binary data (Farris, Kluge, and Eckardt, 97). When the data in a matrix of additive binary characters are clustered using the Wagner algorithm for phylogenetic analysis (Farris, 97; Farris, Kluge, and Eckardt, 97), the Downloaded from at Penn State University (Paterno Lib) on May 2, 26
5 98 HENNIG'S PARASITOLOGICAL METHOD 233 A B C D E F G A b C D E F G 25 original cladogram is always reconstructed. Figures 5 and 6 represent the same parasite and host cladograms shown in Figures 7 and 8, with the internal nodes labeled in Figure 5. Figure 7 depicts the data matrix of converted binary characters based on the parasite data, and Figure 8 shows the host cladogram constructed from the converted data matrix. In this instance, the information-content of the parasite data is completely represented in the host cladogram. Figures 9 and 2 correspond to Figures and 2, with the internal nodes of 9 labeled. The converted data matrix (Fig. 2) and resultant host cladogram (Fig. 22) again depict accurately the host relationships. Thus, in both cases the additive binary coding and cladistic clustering procedure produces the correct set of relationships by representing accurately the available information. The alternative technique of clustering by raw similarity over a set of presence/ absence data will not produce the correct set of relationships under all sets of data because they do not contain or represent all available information. Presence/absence data certainly do not contain the necessary historical components rendering them appropriate for studies of coevolution. Presence/absence data will provide the correct set of relationships under a special array of data, but that is an accident of the array and not a function of the method perse. The additive binary coding and Wagner analysis technique always give the correct representation of the parasite data no matter what the host relationships. This becomes critical when one attempts simultaneous comparisons of two or more parasite taxa with the host cladogram. Downloaded from at Penn State University (Paterno Lib) on May 2, 26 FIGS An example of the technique presented in Figures 5-22 using a real parasite taxon. Figure 23 depicts the cladogram of genera corn- prising the Liolopidae (Digenea) as published by Brooks and Overstreet (978). Figure 24 depicts the relative phylogenetic relationships of the vertebrate taxa hosting liolopids. Figure 25 depicts the Additive Binary Coding matrix for Figure 23. Figure 26 depicts the host cladogram predicted by the parasite data.
6 234 SYSTEMATIC ZOOLOGY VOL. 3 A B C D 27 A B C D E 29 FlCS Misleading estimates of host phylogeny predicted by parasite data resulting from convergent host-parasite relationships. Figures 27 and 28 represent parasite and host taxon cladograms, respectively. Figure 29 depicts the Additive Binary Coding matrix for the parasite cladogram. Based on occurrence of parasite A in hosts I and II, parasite B in hosts III and IV, and parasite C in host V, Figure 3 depicts the phylogenetic relationships of hosts III and IV incorrectly. Figure 3 shows the distribution of parasites among members of the host taxon when host phylogenetic relationships are correctly displayed. Note homoplasious occurrence of parasite B in hosts III and IV. I conceived both examples used so far in such a way that the parasite data are consistent with the host phylogenetic relationships. This is clearly shown by the protocol I espouse, and combining the two data sets would provide only a third congruent cladogram. On the other hand, the raw similarity clustering method tells that one data set indicates perfect co-speciation and the other indicates independent parasites in each host taxon. Combining those two might well provide a third answer, or it might just ignore some information to produce one of the two previously formulated cladograms. There is therefore no reason to use any raw similarity clustering techniques rather than phylogenetic analysis in studies of coevolution. This statement is not meant to be IV IV V V any startling revelation. Studies in coevolution appear to be the one area in which syncretists (Ashlock, 974), pheneticists (Michener, 967) and phylogeneticists (Hennig, 966) have reached accord. Figures demonstrate the method of comparing one parasite cladogram with the host cladogram using the procedure outlined above. The example uses the genera of the digenean family Liolopidae as terminal taxa, based on a cladogram published by Brooks and Overstreet (978). The example parallels Figures 5-8. Up to this point I have considered single parasite terminal taxa or single monophyletic groups in comparison with their host group. The assumption in such cases generally is that the host cladogram is correct; thus, any incongruences exhibited by the parasite data result from convergence rather than from faulty representation of the host phylogenetic relationships. I will show later that this assumption is not necessary. Even with no convergent host-selection, single parasite taxa will not resolve completely or unambiguously host group cladograms unless there has been strict co-speciation. In the absence of convergence, however, the groups depicted as monophyletic by a parasite taxon, even if not completely resolved, will correspond with the host group phylogeny. Comparison of the distribution of members of a single parasite taxon among members of a host group is equivalent to comparing the degree of concordance exhibited by one character (the parasite taxon) with a set of many correlated characters (the host-group cladogram). Morphological homoplasy is detected by phylogeneticists in exactly that manner. This observation allows a reasonable approach to evaluating parasite data. Those parasite groups inhabiting the host group and exhibiting revolutionary relationships would belong to the set of correlated characters depicting the true host phylogeny. Hennig was concerned particularly about parasite data which would give in- Downloaded from at Penn State University (Paterno Lib) on May 2, 26
7 98 HENNIG S PARASITOLOGICAL METHOD 235 correct resolutions of host relationships in the absence of much or any host data, thus negating their usefulness as independent data on host phylogeny. Figure 27 depicts a cladogram for a parasite taxon whose members (A-C) inhabit members of host taxon I-V (Fig. 28). The distribution of parasites is as follows: A inhabits I and II, B inhabits III and IV, and C inhabits V. The converted data matrix is shown in Figure 29 and the resultant host cladogram in Figure 3. Notice that the paraphyletic taxa III and IV have been clustered into a monophyletic taxon. If the host cladogram is available, comparison of host and parasite data will demonstrate that parasite B occurs in III and IV because B is the ancestor of A or because of symplesiomorphous or homoplasious occurrence of B in III and IV (Fig. 3). The second level of using parasite data to reconstruct host phylogenies involves determining a set of correlated parasite characters which are consistent with the host-group phylogeny. The determination of that concordance in the second level of analysis involves comparison of parasite data for a large number of parasite groups simultaneously with the hostgroup cladogram. Most multi-character data sets which are highly correlated with a single cladogram contain characters which do not completely resolve the cladistic relationships of all taxa. Nevertheless, they are all consistent with at least part of the completely resolved cladogram and do not refute any of it. Some characters in multi-character analyses are inconsistent with the most parsimonious cladogram. They owe their incongruity to faulty analysis by the worker or to homoplasy. Yet, as long as they do not represent half or more of the total characters, the incongruent characters will not perturb a cladistic analysis to the point of indicating false relationships. Those properties of multi-character cladistic analyses hold true regardless of type of data used. Therefore, for any parasite fauna of a given host group which includes more than half taxa which occur in their hosts as a result of co-speciation, a cladogram based on analysis of the entire parasite fauna will represent accurately the host group's phylogenetic relationships, for all resolved taxa, and will represent accurately incongruent data. Thus, the problem shown in Figures 27-3 can be overcome. I will show later that one need not assume a priori that more than half the parasite fauna for a given host group exhibits co-speciating relationships with the host-group. Brooks (979b) studied the digeneans infecting crocodilians. He compared cladograms of the various parasite taxa with a cladogram of crocodilian relationships, with patterns of continental drift, and with crocodilian biogeography. That study conforms to my definition of an initial estimate. At that level, the study suggested that crocodilians and their digeneans evolved together as a faunal unit. I used the crocodilian data to prepare a converted data matrix, and formulated a host-group cladogram according to the protocol listed in this paper, choosing the host-group cladogram supported by the predominant pattern of parasite relationships. The data matrix for the analysis contains over binary characters representing 7 transformation series (parasite cladograms). Copies of the parasite cladograms and of the converted data matrix have been deposited along with a hostparasite list for crocodilians and their digeneans (in Brooks, unpublished Ph.D. dissertation) in the Division of Parasitology, University of Nebraska State Museum. Because there is great disparity in numbers of digeneans reported among crocodilians and because there were so many characters in the converted matrix, I had to formulate the crocodilian cladograms in three stages: ) clustering the genera Melanosuchus, Paleosuchus, Osteolaemus, and Gavialis with their sistergenera based on parasites known from those four genera (Figs. 32, 33); 2) clustering the members of Crocodylus by the same criterion (Fig. 34); and 3) clustering Downloaded from at Penn State University (Paterno Lib) on May 2, 26
8 236 SYSTEMATIC ZOOLOGY VOL. 3 Caiman Downloaded from at Penn State University (Paterno Lib) on May 2, FIGS Phylogenetic relationships of crocodilians based on digeneans hosted. Figure 32 depicts the relative relationships of Caiman spp., Melanosuchus and Paleosuchus. Figure 33 depicts relative relationships among Gavialis, Osteolaemus and Crocodylus. Figure 34 depicts relative relationships of Crocodylus spp. Figure 35 depicts relative relationships among Alligator, Caiman and Crocodylus. Figure 36 depicts relative relationships among crocodilian genera for which digeneans have been reported. For further discussion, see text.
9 98 HENNIG S PARASITOLOGICAL METHOD 237 the groups Alligator, Caiman, and Crocodylus (Fig. 35). The hypothesis of cospeciation suggested by Brooks (979b) is corroborated with a single exception. That exception is the placement of Gavialis as the sister-group of Crocodylus- Osteolaemus rather than the sister-group of all living crocodilians (Fig. 36). If among living crocodilians Gavialis truly represents the sister-group of all others, its parasite fauna (only three species are known from it) must include a majority inhabiting Gavialis as a result of ecological convergence with some member(s) of Crocodylus. As with any ambiguous cladogram, more characters are needed to resolve any conflicts in the data set. If the host cladogram is known, it is possible to test whether or not the phylogenetic relationships of the parasites are congruent with host phylogenetic relationships. But what if there is no host cladogram, or if the host cladogram itself is ambiguous? Must parasitologists wait for host-group cladograms to be produced before providing valid information on host-group phylogeny? If so, there is no empirical justification for using parasite data at all. However, such is not the case. A cladogram of host relationships based on parasite data will reflect the pattern of host relationships predicted by the largest number of parasites. Thus, one can examine the internal consistency of the data set simply by observing the degree to which all parasites predict the same set of host relationships. This can be done by finding the shortest unrooted Wagner network (Farris, Kluge, and Eckardt, 97) and then finding the most parsimonious rooted tree from that. I have suggested that a single pattern of host relationships indicated by a correlated set of parasites should be consistent with the pattern of host phylogenetic relationships. That implies conversely that the absence of a highly corroborated pattern should indicate host relationships based on geographic or ecological homoplasy, or on incomplete sampling. Thus, unresolved or ambiguously resolved host relationships indicated by / H Elipesurus MO < 3 ( \ Potamotrygon i \ MA 37 S F H R MO C Y MA FlGS Cladograms depicting phylogenetic relationships among eight species of freshwater stingrays (Potamotrygonidae) based on method outlined in Figures 5-22 and based on data matrix in Appendix. Figure 37 depicts an unrooted Wagner network and Figure 38 a rooted Wagner tree (based on all-plesiomorph ancestor). F = Potamotrygon falkneri; H = P. hystrix; R = P. reticulatus; S = Elipesurus spinicauda; MO = P. motoro; Y = P. yepezi; MA = P. magdalenae. parasite data should alert a systematist that the parasite data may be depicting host phylogenetic relationships incorrectly or ambiguously. The degree of confidence one may place in the phylogenetic relationships of hosts postulated would be determined strictly by the parasite data; other types of data would not be required a priori. In concluding this part, I will present an example of the third level of analysis, that of producing and evaluating a host cladogram in the absence of any data but parasite data. Brooks, Thorson and Mayes (98) recently completed a study in which a parasite fauna, that of Neotropical freshwater stingrays, was analyzed phylogenetically in the absence of any notion of host-group phylogenetic relationships. In that study no attempt was made to formulate a host cladogram, but Downloaded from at Penn State University (Paterno Lib) on May 2, 26
10 238 SYSTEMATIC ZOOLOGY VOL. 3 it was noted that there existed a high degree of consistency among parasite taxa in terms of geographic distribution patterns and host group parasitized by the sister-groups for each parasite taxon analyzed (the second observation is equivalent to using out-group comparisons for determining character-stated polarity). I converted the stingray parasite data into a binary matrix (see Appendix) and formulated cladograms of host relationships from those data (Figs. 37, 38). Calculations for the unrooted and rooted Wagner networks' total homoplasy (Horn option in the Wagner-78 program) produced values of 2 and 22, respectively. Additionally, a number of branches are characterized wholly or in part by the absence of some parasites. These observations clearly announce that there is not a strong pattern indicated for all hosts by the parasite data. Virtually all of the homoplasy is due to parasites found in Potamotrygon reticulatus and P. hystrix. Both stingrays occur sympatrically in the Orinoco River where they share some parasites and exhibit some parasites unique to themselves. The parasites inhabiting Orinocoan stingrays comprise two categories: ) those endemic to the Orinoco area which are related to those inhabiting rays in the Magdalena River and 2) those also occurring in other parts of South America. I believe three factors, two of which involve sampling error, account for the ambiguous data. First, some parasites which have been reported in only one or a few hosts may inhabit others. Secondly, some parasites may occur in more areas than now known. Rhinebothrium paratrygoni and Rhinebothroides scorzai, known from the Orinoco and Parana, probably occur also in the Amazon. Searches for more parasite data could provide evidence clarifying the ambiguity relating to the first two points. Such findings would tend to reduce the homoplasy in the cladograms by placing P. reticulatus and P. hystrix between P. circularis and P. yepezi-p. magdalenae. The third reason for the ambiguity lies in the possibility of convergent host-parasite relationships. If the potamotrygonid stingrays represent a monophyletic grouping, and if geographic and/or ecological isolation played any role in their phylogenesis, the sympatric occurrence of Potamotrygon reticulatus, P. hystrix and Elipesurus spinicauda in the Orinoco River where they share some parasites virtually requires convergent associations. When future work produces data clarifying symplesiomorphic host-parasite relationships and provides enough data to indicate clearly convergent host-parasite relationships, a less ambiguous resolution will be possible. As with any phylogenetic analysis, the answer to ambiguous results is a search for more characters. Of greatest import is the realization that the protocol advocated in this paper produces branching diagrams which point out their own weaknesses. As implied in the last statement, I consider falsification as essential to any empirical method for using parasites as indicators of host phylogeny. Cladograms of host relationships based on parasite data following the approach outlined in this study are exceedingly vulnerable to testing. All one need do is discover new parasites in some hosts or new hosts for some parasites. Additionally, the cladograms of parasite relationships can be modified by complete refutation in some cases or corroborated in others by new data on characters of the parasites themselves. Wiley (975) pointed out that the degree to which any cladistic analysis is corroborated depends on the number and severity of tests applied to it. In one sense, that means the number of new characters discovered which demonstrate the same patterns of relationships as the original cladogram. In another sense, one might consider the cladogram based on the largest number of characters according to the parsimony criterion of accepting the least-refuted cladogram as the best working hypothesis. Pertinent to this study, such reasoning suggests that the degree to which parasites may be viewed as reliable indicators of host phylogenetic relationships depends upon the Downloaded from at Penn State University (Paterno Lib) on May 2, 26
11 98 HENNIG S PARASITOLOGICAL METHOD 239 number of parasite groups examined. This is independent of any a priori assessments of the possibilities of ecological convergence in parasite communities or faunas. A cladogram of host relationships based on all the parasites constituting the parasite fauna of a host group could tell what components of the fauna experienced phylogenesis when the host group did and which did not. Further, such a cladogram would pinpoint elements of the parasite fauna which had co-speciated with the host group and which were invasive, due to host-switching. Additionally, the relative time period during which any invasions occurred would be indicated. In the case of strict co-speciation, this would not yield anything which could not be seen by inspection. However, incorporating such data into the host group cladogram would provide a more informative representation of the available data. PHYLOGENETIC INTERPRETATION OF HOST-PARASITE RELATIONSHIPS As shown in the previous section, any cladogram representing a phylogenetic hypothesis for a parasite taxon may be converted into a matrix of binary characters by additive binary coding. Those characters may then be combined in a branching diagram of host relationships based on the most informative (=parsimonious) representation of correlated parasite data. Once the information-content of many parasite taxa has been presented in terms of hosts, it is possible to determine the phylogenetic context of the postulated host-parasite relationships for each parasite taxon. This is equivalent to comparing a single parasite taxon with a host-group cladogram which was formulated using non-parasite data, but nonparasite data are not required. Those data may, of course, be used when available and may always serve as an additional level of testing. The concern of this paper is the determination of whether or not parasite data could stand by themselves or would always be subsidiary, chorological, data. Parasites exhibit the same kinds of similarity relationships with their hosts as do any other kind of character. This means that any single parasite taxon, like any other single character, may not completely resolve the phylogenetic relationships of its host group and may give misleading results if there are homoplasious or symplesiomorphous host-parasite relationships (Figs. 27-3). These problems may be overcome by increasing the number of parasite taxa considered if a consistent pattern of host relationships emerges. This is not based on an assumption that most host-parasite relationships result from co-speciation; rather, it is based on the observation that a single consistent pattern of host relationships indicated by the parasite data is inconsistent with the alternative to co-speciation, namely, random colonization or host-switching. I will consider later the possibility of a special case of host-switching in which host-switching imitates co-speciation. The greater the internal consistency of the parasite data, the greater the degree of confidence one may have that the host relationships postulated are their phylogenetic relationships. Before discussing the phylogenetic interpretation of hostparasite relationships postulated by an analysis such as I have suggested in the previous section, I must point out that the analytical basis of this study is independent of any models, theories, or hypotheses about coevolution. The utility of using parasite data in delineating host phylogenetic relationships is not affected by the processes whereby the parasites became associated with particular hosts. However, for evolutionary biologists interested in the historical associations of hosts and parasites, such distinctions may be critical. For example, the occurrence of parasite B in hosts III and IV in Figures 27-3 may be the result of homoplasious or of symplesiomorphous host-parasite relationships. A priori determination of which is not required for analysis of patterns of host relationships indicated Downloaded from at Penn State University (Paterno Lib) on May 2, 26
12 24 SYSTEMATIC ZOOLOGY VOL. 3 by parasite data because in either event the same branching diagram would be produced. This is because the analysis is based on a search for the most parsimonious arrangement of observations and the determination of symplesiomorphy or parallelism is a phylogenetic inference from the analysis, made a posteriori. Postulated host-parasite relationships may represent one of four general types: synapomorphous, symplesiomorphous, convergent, or parallel. Each of those may corroborate more than one phylogenetic interpretation, listed below. Synapomorphous host-parasite relationships represent episodes of strict co-speciation. This means that host and parasite phylogenies are maximally correlated. In the next section, I will examine contrasting revolutionary scenarios, one in which parasite phylogenesis is condered a dependent variable on evolutionary changes of independently associated hosts, and one in which both host and parasite phylogenesis are considered independent variables. Each model explains co-speciation differently. Convergent host-parasite relationships are those which appear as single or randomly distributed autapomorphies of hosts. They are de facto evidence of host transfers, or broadening co-accommodation. They are necessary but not sufficient observations for supporting a coevolutionary scenario involving sympatric speciation; this will be discussed in the next section. Symplesiomorphous and parallel hostparasite relationships will be considered together both because they are indistinguishable at the analytical level and because their occurrence provides critical data for evaluating various models of coevolution. Symplesiomorphous host-parasite relationships comprise two general types, each of which is consistent with a different model of co-evolution. The first type is exemplified by the example shown in Figures 39-4, wherein four parasite taxa inhabit five host taxa. The most parsimonious phylogenetic interpretation of the occurence of parasite B in hosts II and III is that the host group underwent two episodes of phylogenesis during which parasite B remained with all descendant hosts and did not experience phylogenesis itself. Such an interpretation is consistent with both host and parasite cladograms and requires no additional conditions or assumptions. Such an occurrence could be the result of parallelism but, as I will show later, that would require at least one extra assumption. The second type of symplesiomorphic host-parasite relationship is reflected in Figures In this case, the occurrence of parasite B in hosts III and IV could be symplesiomorphous rather than homoplasious only if one phylogenetic interpretation and two analytical conditions were met. The phylogenetic interpretation is that parasite B would be the ancestor of parasite A. The conditions which must be met include ) B must exhibit a branch length of zero in any cladogram of parasite relationships and 2) III must not host parasite A. Assuming that B is the ancestor of A produces a new tree (Fig. 4) and converted data matrix (Fig. 42). The resultant host cladogram (Fig. 43) does not identify III and IV as a monophyletic group, but neither does it correctly place III and IV in their paraphyletic sequence. It is not exactly wrong, but it is not correct, either. Correct resolution would require at least one additional character whether B was considered an ancestor or a sister-taxon, so a priori designation of ancestors is not necessary for proper representation of host relationships at the analytical level. Treating some terminal taxa as ancestors at the analytical level is not feasible for a number of reasons. First, it is difficult to defend the position that one extant taxon is the ancestor of another extant taxon, especially if the parasite to be designated the ancestor exhibits a branch length greater than zero in a cladistic analysis. In such a case, depiction of one taxon as ancestor would not be the most parsimonious representation of the available data, and would introduce unacceptable bias. Secondly, even if one tax- Downloaded from at Penn State University (Paterno Lib) on May 2, 26
13 98 HENNIG S PARASITOLOGICAL METHOD 24 (A) (B) II (B) (C) IV (D) V 39 4 FlGS Symplesiomorphous occurrence of a parasite taxon in two host taxa. Figure 39 shows host-group cladogram for taxa I-V with associated parasites A-D above. Figure 4 shows cladogram for parasite taxon A-D. Occurrence of B in II and III is most parsimoniously attributed to symplesiomorphy, wherein host group underwent two episodes of phylogenesis during which parasite group underwent none, but remained associated with all descendants. For fuller explanation, see text. on exhibits a branch length of zero, it may not be the ancestor of another included in the analysis. Thus, there are no independent a priori criteria for considering any terminal taxon the ancestor of another. The choice of depicting B as the ancestor of A was based solely on a desire to fit the parasite data to the host cladogram even if it meant altering the results A B C D A B C D 42 Downloaded from at Penn State University (Paterno Lib) on May 2, 26 FIGS Estimating host phylogeny if some parasites are considered ancestors of other parasites among living taxa. Figure 4 depicts the parasite taxon from Figure 27 with terminal taxon B considered ancestral to A. Figure 42 depicts the Additive Binary Coding matrix for Figure 4. Figure 43 depicts the resulting cladogram based on the same D distribution of parasite among hosts as listed for Figures Figure 44 depicts host relationships predicted by parasite data if A is considered ancestral to B in Figure 27 or 32. Correct host phylogenetic relationships are presented in Figure 28.
14 242 SYSTEMATIC ZOOLOGY VOL. 3 of the cladistic analysis. We could just as easily have considered A the ancestor of B and the resulting cladogram (Fig. 44) would be even worse than the original (Fig. 43) which at least depicted I and II as a monophyletic group. And thirdly, treating one terminal taxon as the ancestor of another has the effect in an analytical procedure of simply reducing the effect of a problematical character-state; hence the ambiguous results of Figure 43. It is a means of ignoring inconvenient data. Absolutely any notion of coevolutionary relationships could be supported by ad hoc invocation of an ancestor-descendant relationship. And finally, adopting such a convention would require two assumptions: ) the host-group cladogram is always correct and 2) occurrence of the same parasite in two or more paraphyletic taxa is never the result of ecological homoplasy but always the result of co-speciation. I view both of those assumptions as being contrary to the aims of an empirical method in phylogenetics. Designations of ancestor-descendant relationships therefore cannot be assumed a priori and can be supported only if the conditions listed above are met as the result of an analysis of the type suggested in this study. Both types of host-parasite relationships discussed in the previous section could be the result of homoplasy due to host transfer. In the second case (Figs. 27-3), if parasite B has a branch length greater than zero and III and IV are sympatric or parapatric, B cannot be considered the ancestor of A under any circumstances and the occurrence of B in III and IV must be due to homoplasy. If III also hosts A, the hypothesis of parallelism is corroborated without additional assumptions. If III hosts only B it must be assumed that the phylogenetic episode which produced host III and the ancestor of I + II was accompanied by the loss of any member of the parasite lineage in III, followed by parallel acquisition of parasite B at a later time. For the first type discussed above (Figs. 39-4) this is a less parsimonious statement than an interpretation of symplesiomorphy; for the second type, parallelism is more parsimonious than an interpretation of symplesiomorphy based on presumed ancestor-descendant relationships. Thus, for data not directly referrable to co-speciation (synapomorphy) or random colonization via host-switching (convergence), symplesiomorphic occurrence of sistergroups and parallel occurrence of a taxon are more parsimonious interpretations than symplesiomorphic occurrence of a persistent ancestor. COMPARING COEVOLUTIONARY MODELS Most models of coevolution reflect one of two general viewpoints. First, parasite phylogenesis may be a colonization phenomenon, dependent upon ability to switch hosts, often equated with degree of co-accommodation, or upon host phylogenesis. Secondly, parasite phylogenesis may be independent of degree of coaccommodation and generally coincides with host phylogenesis because hosts and parasites experience the same allopatric speciation events. In the first model, congruent host and parasite phylogenies are predicted only when coaccommodation is very narrow, and not always then (e.g., Bush, 975), a condition not encountered often enough to serve as a general rule. Under such a model, parasites could be related to each other in a manner consistent with the phylogenetic relationships of their hosts, but no members of the parasite group would ever have infected some ancestral hosts. Such a non-random colonization scenario is embodied in the random colonization or "resource tracking" models of coevolution and to an extent in some models of parasite sympatric speciation (e.g., Bush, 975), wherein congruent host and parasite phylogenies are viewed as occasional outcomes of a random process usually involving spatially proximate but phylogenetically distant hosts. The colonization type of mechanism could produce patterns imitating co-speciation (Fig. 45). An ancestral parasite was acquired either through host-switch- Downloaded from at Penn State University (Paterno Lib) on May 2, 26
15 98 HENNIG S PARASITOLOGICAL METHOD 243 (I) 45a IV II IV III IV 47a 47b FlGS Phylogenetic implications of "resource-tracking" models of coevolution. Figure 45 depicts two variations (45a and 45b) of the process involved in "phylogenetic tracking." Figure 46 depicts a cladogram of parasite relationships for the four parasites listed in Figure 45; Roman numerals are parasites, letters are hosts. Figure 47a depicts the correct phylogenetic relationships for the four parasite taxa shown in Figure 45a and Figure 47b depicts the correct phylogenetic relationships for the four parasite taxa shown in Figure 45b if phylogenetic resource tracking protocol actually accounts for observed hostparasite relationships. For a fuller discussion, refer to text. ing from a host outside the study host group or inhabited the common ancestor of the study host group (Fig. 45a). When host phylogenesis occurred, one descendant (D) retained the parasite and another (B) lost it or D acquired the parasite from a host outside the study host group (X in Fig. 45a). Host B then experienced phylogenesis, resulting in descendants C and E. Then, parasites inhabiting D (I in Fig. 45) colonized E and may or may not have colonized C. Subsequently, the members of I inhabiting E experienced phylogenesis, becoming parasite taxon II. The process then repeated until the phylogeny of the hosts shown in Figure 45 was complete and the parasites shown as inhabiting those hosts had become distinct entities. I have two reservations about the above scenario. First, the observation that parasites "track" or require certain host resources does not make the above scenario any more likely than any other. In fact, resource-tracking in parasites does not provide any information about Downloaded from at Penn State University (Paterno Lib) on May 2, 26
16 244 SYSTEMATIC ZOOLOGY VOL. 3 parasite phylogenesis. Both models under scrutiny require resource-tracking, which is a manifestation of the aspect of coevolution termed co-accommodation (Brooks, 979a) and thus is not directly relevant to the problem of using parasite data for reconstructing host phylogenies or evaluating coevolutionary scenarios. Secondly, there are a number of assumptions which must be adopted by proponents of this model. These assumptions are not expressed explicitly for the most part, but they are important nonetheless. The required assumptions comprise three categories. First, there is a set of assumptions concerning the relative time of appearance for each host and each parasite taxon. The model asserts that each host taxon undergoes phylogenesis without the parasite group and exists as a distinct entity before it acquires the parasite. The parasite colonizers then experience phylogenesis. Thus, parasite phylogenesis proceeds more slowly than host phylogenesis; the parasite taxa must "wait" for new host taxa to be produced. A further assumption requires that some host descendants either lose their parasites or never have parasites even though their sister-taxa and descendants are parasitized. Because any descendants can become ancestors themselves and because all ancestors were descendants at some time, such a model places an investigator in the position of trying to defend a logical paradox. The second set of assumptions concerns the geographical and/or ecological relationships of the hosts. This model requires some form of isolation (ecological, geographical, or both) of hosts, followed by a breakdown in that isolation to the extent that parasite colonization occurs, followed further by renewed isolation such that parasite phylogenesis occurs. For hosts and parasites exhibiting congruent cladograms and occurring allopatrically, such assumptions are not defensible. It is not beyond the realm of possibility for such an isolation-sympatry-isolation scenario to occur at some point. However, it seems unlikely that ) such occurrences could have existed consistently for a parasite lineage, that 2) they would always conform to the phylogenetic relationships of the hosts, or that 3) an entire parasite fauna would exhibit phylogenetic relationships congruent with host relationships by such means. Assumptions concerning the phylogenetic relationships of the parasite taxa comprise the third set of assumptions required by the "tracking" model. Simply put, this model requires that some extant parasites be ancestors of other extant parasites at any given time. At the analytical level, it will require that some known extant parasites always be designated as ancestors of other known parasites. Consider the cladogram in Figure 46 for the parasites occurring in hosts shown in Figure 45. For episodes of co-speciation, nodes 5, 6, and 7 are defined they represent parasite taxa occurring in the common ancestors C, B, and A (Fig. 45), respectively. For non-random colonization, 5, 6, and 7 are undefined without referring to extant taxa already known (Fig. 47). If the branches of the cladogram in Figure 46 all include apomorphic traits, the representations in Figure 47 are not possible. Thus, a "tracking" model postulates that in general there would not be a high degree of concordance between host and parasite cladograms. Further, such a model asserts that for cases in which such concordance occurs parasites must exist in geographic distribution patterns not referable to vicariance and must satisfy the above assumptions. Such a model apparently is based on an assumption that parasites evolve more slowly than their hosts. Szidat (956) espoused such an idea based on the notion that parasites inhabited more stable niches than did free-living organisms. Price (977) discussed some general properties of the evolutionary biology of parasites and concluded that parasites exhibit very high speciation rates. That was based on the observation that there are more parasitic than free-living taxa in ex- Downloaded from at Penn State University (Paterno Lib) on May 2, 26
17 98 HENNIG S PARASITOLOGICAL METHOD 245 istence. Szidat's assertion (see also Stammer, 957) also assumes that the environmental variations experienced by a host are not reflected in the host's physiology and homeostasis. Evolutionary, such a belief requires that part of any host organism (the organs associated with the parasites' habitats) evolves more slowly than the rest of the host. I will not pursue this point's absurdity except to point out that Mitchell (9) was able to construct an account of avian phylogeny based on intestinal modifications, suggesting that that rich habitat for parasites evolved right along with the rest of the body. While it is entirely possible that parasites evolve more slowly than hosts, at least in some cases, I cannot embrace any methodology which requires such an assumption at the outset. I will show next that there is a more parsimonious model which explains our observations. Regenfuss (978) noted that host and parasite phylogenies often have elements which parallel each other. He suggested that Hennigian analysis of host and of parasite groups was necessary for best documentation of such occurrences. Regenfuss further stated that parallel phylogenesis of host and parasite taxa always occurs in close conjunction with parasite transfers, and that close consideration should be given to the time of initial colonization of the host group and of the parasite transfer. I have shown that a model of coevolution based on host transfers requires that some extant parasite species always be ancestors of other extant species. If Hennigian analysis, which recognizes only sister-group relationships, is necessary to document congruent host and parasite phylogenesis, host transfer cannot be the causal agent of the congruence. Furthermore, it is not entirely clear how workers are to determine the time of colonization and of host transfers in coevolution before performing some sort of phylogenetic analysis. Alternatively to the "tracking" models, parasitologists studying parasite systematics have noted for a long time that most parasite groups exhibit non-random associations with host groups. Many parasitologists have suspected also that vicariant geographical distributions of closely related parasite taxa did not result from long-range dispersal in a random manner but from historical alterations in geography. Indeed, when host and parasite cladograms are compared there is usually a high degree of concordance between the two. And when parasite and geographic cladograms are compared, the concordance is even more striking. Brooks (979a) accounted for this general pattern of concordance by referring to the allopatric speciation/vicariance biogeography model, suggesting that hosts and parasites exhibited a general pattern of congruent phylogenies because their ancestors experienced the same allopatric speciation events. Brooks further suggested that such a viewpoint was consistent with the views of parasite systematists beginning with von Ihering (89) and espoused recently by Inglis (97). Coincidence of a consistent set of parasite relationships with vicariant geographical distributions, such that parasites and geography have coevolved, would support an hypothesis of host-parasite co-speciation. The alternative would be that the parasites had evolved without their current hosts; that is, they evolved in one set of hosts, then colonized randomly their present hosts while their original hosts either became extinct or simply lost their original parasite fauna. Such a scenario seems rather unlikely for even a single parasite taxon, but approaches near impossibility when an entire parasite fauna is considered. For example, the cladistic relationships of crocodilian digeneans corresponded to vicariant geographical distribution patterns consistent among the entire set of parasite taxa and also consistent with the biogeographical relationships of the host group and with patterns of continental drift. Upon checking the host genealogy, I discovered a high degree of concordance remained. Thus, it was not surprising that the parasite-data cladogram of crocodilian relationships Downloaded from at Penn State University (Paterno Lib) on May 2, 26
18 246 SYSTEMATIC ZOOLOGY VOL. 3 (A) (B) (C) I II III (Y) (X) (A) (B) (C) (D) IV (Z) (D) IV FiGS Demonstration that degree of coaccommodation (host specificity) need not be great in order for the method proposed in this study to be matched well with a cladogram based on crocodilian morphology. At first thought it might seem that my method is biased towards producing spurious co-speciation patterns and supporting the vicariance model because co-accommodation is not considered relevant to the analytical method. Holmes and Price (98) presented evidence concerning co-accommodation in responding to my observations about the co-speciation of hosts and parasite communities (Brooks, 98). Thus, there is a perception that co-speciation is just an occasional result of variations in co-accommodation. The final set of figures demonstrates that my method, if biased at all, will be biased against co-speciating relationships. Consider monophyletic host-group A-D (Fig. 48) and parasite group I-IV (Fig. 49). Initially, the parasite group exhibits co-speciation with the host group. If, in the course of time, some of the parasites begin inhabiting other hosts outside the group A-D (Fig. 5), broadening their co-accommodation, the co-speciation message still remains discernible using my method. Thus, different degrees of co-accommodation are not relevant. If for some reason the original hosts (B and D in Fig. 49) no longer host the parasites which originally co-speciated with them, as shown in Figure 5, the method presented herein would stipulate convergent rather than co-speciating host-parasite relationships. Thus, if there is any valid, and that disregarding co-accommodation will never result in over-estimation of co-speciation patterns but rather to underestimation of such patterns. Figure 48 depicts a host taxon's phylogenetic relationships. Figure 49 shows that the parasite taxon I II III IV has co-speciated with the host taxon. Figure 5 depicts broadening co-accommodation by some members of the parasite taxon; note that cospeciation of ABCD and I-II III-IV can still be discerned. Figure 5 depicts the host-parasite relationships of parasite taxon I II III IV if B and D no longer host members of the parasite taxon. In this case, even though co-speciation occurred, the host-parasite relationships would be considered convergent just like any other truly convergent host-parasite relationships. Downloaded from at Penn State University (Paterno Lib) on May 2, 26
19 98 HENNIG S PARASITOLOGICAL METHOD 247 bias inherent in the method proposed in this paper, it is such that it precludes some co-speciating taxa from being recognized rather than postulating spurious co-speciating relationships. The reason this happens is that the method is based on observed rather than assumed hostparasite relationships. It sacrifices possible intuitive insights in favor of a method which does not require any a priori assumptions either of co-speciation or of random colonization in producing hostparasite reaationships. CONCLUSIONS What can biologists gain from parasite data used in phylogenetic studies of freeliving organisms? Students of the host group undoubtedly would be interested in knowing which parasites exhibited any of the postulated phylogenetic relationships with their hosts; otherwise, some parasites could be used to support a syncretistic or artificial classification system. Additionally, because parasites are excellent ecological indicators (especially those with complex life cycles), their phylogenetic relationships with the host group provide clues to the ecological evolution of the group. Parasitologists would like to know about the coevolutionary relationships among hosts and parasites because parasite survival is always dependent on some host, and it is interesting to investigate how parasite groups have managed to survive in the same host lineage despite host phylogenesis, or in different host groups, which requires some form of host-switching. Secondary acquisition or secondary loss of a parasite taxon provides grounds for studying the historical development of the host group's ecological and immunological traits. Discovery of immunological traits which confer resistance or immunity undoubtedly would be enhanced if comparisons were made between sister-taxa, one immune or resistant and one susceptible, rather than between model organism and target organism which are not closely related. Likewise, the search for examples of competitive exclusion or of random colonization in community or faunal structures requires knowledge of historical relationships of parasites infecting a given host group (Brooks, 98). Thus, comparison of a parasite taxon's cladogram with a host group's cladogram may indicate interesting relationships worthy of future study by ecologists, immunologists and epidemiologists as well as by phylogeneticists. Comparison of many cladograms for members of parasite faunas with the cladogram of host relationships may provide information for ecologists and evolutionary biologists regarding phylogenetic constraints on or phylogenetic components of community or faunal structure. Such studies may provide means of adopting restrictive (informative) hypotheses of evolutionary processes producing contemporary community or faunal structure. And finally, in the absence of a formulated host cladogram a coevolving parasite fauna may provide data adequate for producing a cladogram of host phylogenetic relationships. Parasitological data constitute a rich potential source of phylogenetic information. Hennig's fears that reliance upon parasitological data would result in the formation of many para- and polyphyletic taxa as groups were misapprehensions. Such potential problems are no greater with parasite data than with any other characters, and in any event can be circumscribed by analyzing a large number of taxa simultaneously and using a parsimony method of phylogenetic analysis to determine the presence and form of any consistent pattern in the data. Formulating parasite cladograms and comparing their concordance with the patterns of historical biogeographic relationships as well as with the hostgroup cladogram represents the best method currently available for providing useful information concerning any single group of parasites for biologists working with free-living organisms. The analytical method described in this paper provides a means of analyzing large numbers Downloaded from at Penn State University (Paterno Lib) on May 2, 26
20 248 SYSTEMATIC ZOOLOGY VOL. 3 of parasite taxa simultaneously, and provides one solution to Hennig's Parasitological Method dilemma. ACKNOWLEDGMENTS Two post-hennig developments in phylogenetic systematics allowed the above formalization. They are related because they were formulated primarily by the same person, James S. Farris, State University of New York at Stony Brook. They are also related because they allow parasite data to be incorporated into a general reference system without any loss of information. Those two developments are the Additive Binary Coding technique and the Wagner Analysis algorithm. Parasitologists in particular and students of coevolution in general owe Dr. Farris a debt of gratitude. I owe him a personal debt of thanks for his unselfish advice, tutelage, and friendship. My thanks also to those who have encouraged my efforts in studying parasite phylogenetics: John D. Lynch, Gareth Nelson, Norman I. Platnick, Randall T. Schuh, and James D. Smith. Jack Frazier, Vicki Funk, Larry Watrous, and Quentin Wheeler participated in many spirited discussions. Mss. Sigrid James and Janine Caira prepared the illustrations. This study is dedicated to Lucien Douglas Brooks and Duncan McVeigh Brooks, deceased. REFERENCES ASHLOCK, P. D The uses of cladistics. Ann. Rev. Ecol. Syst, 5:8-99. BROOKS, D. R Evolutionary history of some plagiorchioid trematodes of anurans. Syst. Zool., 26: BROOKS, D. R. 978a. Systematic status of the proteocephalid cestodes of North American reptiles and amphibians. Proc. Helminthol. Soc. Washington, 45:-28. BROOKS, D. R. 978b. Evolutionary history of the cestode order Proteocephalidea. Syst. Zool., 27: BROOKS, D. R. 979a. Testing the context and extent of host-parasite coevolution. Syst. Zool., 28: BROOKS, D. R. 979b. Testing hypotheses of evolutionary relationships among parasitic helminths: The digeneans of crocodilians. Amer. Zool., 9: BROOKS, D. R. 98. Allopatric speciation and noninteractive parasite community structure. Syst. Zool., 29: BROOKS, D. R. 98a. Revision of the Acanthostominae (Digenea: Cryptogonimidae). Zool. Proc. Linnean Soc, 7: BROOKS, D. R., M. A. MAYES, AND T. B. THORSON. 98b. Systematic review of cestodes infecting freshwater stingrays (Chondrichthyes: Potamotrygonidae) including four new species from Venezuela. Proc. Helminthol. Soc. Washington, 48: BROOKS, D. R., AND R. M. OVERSTREET The family Liolopidae (Digenea), including a new genus and two new species from crocodilians. Int. J. Parasitol., 8: BROOKS, D. R., T. B. THORSON, AND M. A. MAYES. 98. Freshwater stingrays (Potamotrygonidae) and their helminth parasites: Testing hypotheses of evolution and coevolution, in Advances in Cladistics: Proceedings of the First Meeting of the Willi Hennig Society (V. A. Funk and D. R. Brooks, eds.). New York Botanical Gardens, New York, in press. BUSH, G. L Sympatric speciation in phytophagous parasitic insects. Pp. 8-26, in Evolutionary Strategies of Parasitic Insects and Mites (P. W. Price, ed.). Plenum Press, New York, 224 pp. DEARDORFF, T. L., D. R. BROOKS, AND T. B. THOR- SON. 98. Two species of Echinocephalus (Nematoda: Gnathostomidae) from neotropical stingrays. J. Parasitol., 67:in press. FARRIS, J. S. 97. Methods for computing Wagner trees. Syst. Zool., 9: FARRIS, J. S Formal definitions of paraphyly and polyphyly. Syst. Zool., 23: FARRIS, J. S., A. G. KLUGE, AND M. J. ECKARDT. 97. A numerical approach to phylogenetic systematics. Syst. Zool., 9: HENNIG, W Phylogenetic Systematics. Univ. Illinois Press, Urbana, 263 pp. HOLMES, J. C, AND P. W. PRICE. 98. Parasite communities: The roles of phylogeny and ecology. Syst. Zool., 29: IHERING, H. VON. 89. On the ancient relations between New Zealand and South America. Proc. New Zealand Inst., 24: KETHLEY, J. B., AND D. E. JOHNSTON Resource tracking patterns in bird and mammal ectoparasites. Misc. Publ. Ent. Soc. America, 9: MANTER, H. W Parasites of fishes as biological indicators of recent and ancient conditions. Pp. 59-7, in Proc. 26th Oregon State Univ. Biol. Colloq. (J. E. McCauley, ed.). Oregon State Univ. Press, Corvallis, 48 pp. MICHENER, C. D Diverse approaches to systematics. Pp. -38, in Evolutionary Biology, (T. Dobzhansky, ed.). Appleton-Century-Crofts, New York. MITCHELL, P. C. 9. On the intestinal tract of birds; with remarks on the valuation and nomenclature of zoological characters. Trans. Linnean Soc, London, 8: PRICE, P. W General concepts on the evolutionary biology of parasites. Evolution, 3: REGENFUSS, H [Causes and consequences of a parallel phylogenetic cleavage of parasites and hosts.] Sonderb. Naturwiss. Ver. Hamburg, 2:83-. STAMMER, H. J Gedanken zu den parasitophyletischen Regeln und zur Evolution der Parasiten. Zool. Anzeig., 59: Downloaded from at Penn State University (Paterno Lib) on May 2, 26
Phylogeny Reconstruction
 Phylogeny Reconstruction Trees, Methods and Characters Reading: Gregory, 2008. Understanding Evolutionary Trees (Polly, 2006) Lab tomorrow Meet in Geology GY522 Bring computers if you have them (they will
Phylogeny Reconstruction Trees, Methods and Characters Reading: Gregory, 2008. Understanding Evolutionary Trees (Polly, 2006) Lab tomorrow Meet in Geology GY522 Bring computers if you have them (they will
HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS
 HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS WASHINGTON AND LONDON 995 by the Smithsonian Institution All rights reserved
HAWAIIAN BIOGEOGRAPHY EVOLUTION ON A HOT SPOT ARCHIPELAGO EDITED BY WARREN L. WAGNER AND V. A. FUNK SMITHSONIAN INSTITUTION PRESS WASHINGTON AND LONDON 995 by the Smithsonian Institution All rights reserved
Bio 1B Lecture Outline (please print and bring along) Fall, 2006
 Bio 1B Lecture Outline (please print and bring along) Fall, 2006 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #4 -- Phylogenetic Analysis (Cladistics) -- Oct.
Bio 1B Lecture Outline (please print and bring along) Fall, 2006 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #4 -- Phylogenetic Analysis (Cladistics) -- Oct.
History of Lineages. Chapter 11. Jamie Oaks 1. April 11, Kincaid Hall 524. c 2007 Boris Kulikov boris-kulikov.blogspot.
 History of Lineages Chapter 11 Jamie Oaks 1 1 Kincaid Hall 524 joaks1@gmail.com April 11, 2014 c 2007 Boris Kulikov boris-kulikov.blogspot.com History of Lineages J. Oaks, University of Washington 1/46
History of Lineages Chapter 11 Jamie Oaks 1 1 Kincaid Hall 524 joaks1@gmail.com April 11, 2014 c 2007 Boris Kulikov boris-kulikov.blogspot.com History of Lineages J. Oaks, University of Washington 1/46
Introduction to Cladistic Analysis
 3.0 Copyright 2008 by Department of Integrative Biology, University of California-Berkeley Introduction to Cladistic Analysis tunicate lamprey Cladoselache trout lungfish frog four jaws swimbladder or
3.0 Copyright 2008 by Department of Integrative Biology, University of California-Berkeley Introduction to Cladistic Analysis tunicate lamprey Cladoselache trout lungfish frog four jaws swimbladder or
Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes)
 Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes) Phylogenetics is the study of the relationships of organisms to each other.
Introduction to phylogenetic trees and tree-thinking Copyright 2005, D. A. Baum (Free use for non-commercial educational pruposes) Phylogenetics is the study of the relationships of organisms to each other.
Title: Phylogenetic Methods and Vertebrate Phylogeny
 Title: Phylogenetic Methods and Vertebrate Phylogeny Central Question: How can evolutionary relationships be determined objectively? Sub-questions: 1. What affect does the selection of the outgroup have
Title: Phylogenetic Methods and Vertebrate Phylogeny Central Question: How can evolutionary relationships be determined objectively? Sub-questions: 1. What affect does the selection of the outgroup have
muscles (enhancing biting strength). Possible states: none, one, or two.
 Reconstructing Evolutionary Relationships S-1 Practice Exercise: Phylogeny of Terrestrial Vertebrates In this example we will construct a phylogenetic hypothesis of the relationships between seven taxa
Reconstructing Evolutionary Relationships S-1 Practice Exercise: Phylogeny of Terrestrial Vertebrates In this example we will construct a phylogenetic hypothesis of the relationships between seven taxa
LABORATORY EXERCISE 6: CLADISTICS I
 Biology 4415/5415 Evolution LABORATORY EXERCISE 6: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Biology 4415/5415 Evolution LABORATORY EXERCISE 6: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
LABORATORY EXERCISE 7: CLADISTICS I
 Biology 4415/5415 Evolution LABORATORY EXERCISE 7: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
Biology 4415/5415 Evolution LABORATORY EXERCISE 7: CLADISTICS I Take a group of organisms. Let s use five: a lungfish, a frog, a crocodile, a flamingo, and a human. How to reconstruct their relationships?
1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters
 1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. The sister group of J. K b. The sister group
1 EEB 2245/2245W Spring 2014: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. The sister group of J. K b. The sister group
Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1
 Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1 Systematics is the comparative study of biological diversity with the intent of determining the relationships between organisms. Humankind has always
Geo 302D: Age of Dinosaurs LAB 4: Systematics Part 1 Systematics is the comparative study of biological diversity with the intent of determining the relationships between organisms. Humankind has always
CLADISTICS Student Packet SUMMARY Phylogeny Phylogenetic trees/cladograms
 CLADISTICS Student Packet SUMMARY PHYLOGENETIC TREES AND CLADOGRAMS ARE MODELS OF EVOLUTIONARY HISTORY THAT CAN BE TESTED Phylogeny is the history of descent of organisms from their common ancestor. Phylogenetic
CLADISTICS Student Packet SUMMARY PHYLOGENETIC TREES AND CLADOGRAMS ARE MODELS OF EVOLUTIONARY HISTORY THAT CAN BE TESTED Phylogeny is the history of descent of organisms from their common ancestor. Phylogenetic
INQUIRY & INVESTIGATION
 INQUIRY & INVESTIGTION Phylogenies & Tree-Thinking D VID. UM SUSN OFFNER character a trait or feature that varies among a set of taxa (e.g., hair color) character-state a variant of a character that occurs
INQUIRY & INVESTIGTION Phylogenies & Tree-Thinking D VID. UM SUSN OFFNER character a trait or feature that varies among a set of taxa (e.g., hair color) character-state a variant of a character that occurs
Species: Panthera pardus Genus: Panthera Family: Felidae Order: Carnivora Class: Mammalia Phylum: Chordata
 CHAPTER 6: PHYLOGENY AND THE TREE OF LIFE AP Biology 3 PHYLOGENY AND SYSTEMATICS Phylogeny - evolutionary history of a species or group of related species Systematics - analytical approach to understanding
CHAPTER 6: PHYLOGENY AND THE TREE OF LIFE AP Biology 3 PHYLOGENY AND SYSTEMATICS Phylogeny - evolutionary history of a species or group of related species Systematics - analytical approach to understanding
Modern Evolutionary Classification. Lesson Overview. Lesson Overview Modern Evolutionary Classification
 Lesson Overview 18.2 Modern Evolutionary Classification THINK ABOUT IT Darwin s ideas about a tree of life suggested a new way to classify organisms not just based on similarities and differences, but
Lesson Overview 18.2 Modern Evolutionary Classification THINK ABOUT IT Darwin s ideas about a tree of life suggested a new way to classify organisms not just based on similarities and differences, but
Cladistics (reading and making of cladograms)
 Cladistics (reading and making of cladograms) Definitions Systematics The branch of biological sciences concerned with classifying organisms Taxon (pl: taxa) Any unit of biological diversity (eg. Animalia,
Cladistics (reading and making of cladograms) Definitions Systematics The branch of biological sciences concerned with classifying organisms Taxon (pl: taxa) Any unit of biological diversity (eg. Animalia,
What are taxonomy, classification, and systematics?
 Topic 2: Comparative Method o Taxonomy, classification, systematics o Importance of phylogenies o A closer look at systematics o Some key concepts o Parts of a cladogram o Groups and characters o Homology
Topic 2: Comparative Method o Taxonomy, classification, systematics o Importance of phylogenies o A closer look at systematics o Some key concepts o Parts of a cladogram o Groups and characters o Homology
Lecture 11 Wednesday, September 19, 2012
 Lecture 11 Wednesday, September 19, 2012 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean
Lecture 11 Wednesday, September 19, 2012 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean
UNIT III A. Descent with Modification(Ch19) B. Phylogeny (Ch20) C. Evolution of Populations (Ch21) D. Origin of Species or Speciation (Ch22)
 UNIT III A. Descent with Modification(Ch9) B. Phylogeny (Ch2) C. Evolution of Populations (Ch2) D. Origin of Species or Speciation (Ch22) Classification in broad term simply means putting things in classes
UNIT III A. Descent with Modification(Ch9) B. Phylogeny (Ch2) C. Evolution of Populations (Ch2) D. Origin of Species or Speciation (Ch22) Classification in broad term simply means putting things in classes
1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters
 1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. Identify the taxon (or taxa if there is more
1 EEB 2245/2245W Spring 2017: exercises working with phylogenetic trees and characters 1. Answer questions a through i below using the tree provided below. a. Identify the taxon (or taxa if there is more
Systematics, Taxonomy and Conservation. Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem
 Systematics, Taxonomy and Conservation Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem What is expected of you? Part I: develop and print the cladogram there
Systematics, Taxonomy and Conservation Part I: Build a phylogenetic tree Part II: Apply a phylogenetic tree to a conservation problem What is expected of you? Part I: develop and print the cladogram there
Fig Phylogeny & Systematics
 Fig. 26- Phylogeny & Systematics Tree of Life phylogenetic relationship for 3 clades (http://evolution.berkeley.edu Fig. 26-2 Phylogenetic tree Figure 26.3 Taxonomy Taxon Carolus Linnaeus Species: Panthera
Fig. 26- Phylogeny & Systematics Tree of Life phylogenetic relationship for 3 clades (http://evolution.berkeley.edu Fig. 26-2 Phylogenetic tree Figure 26.3 Taxonomy Taxon Carolus Linnaeus Species: Panthera
Inferring Ancestor-Descendant Relationships in the Fossil Record
 Inferring Ancestor-Descendant Relationships in the Fossil Record (With Statistics) David Bapst, Melanie Hopkins, April Wright, Nick Matzke & Graeme Lloyd GSA 2016 T151 Wednesday Sept 28 th, 9:15 AM Feel
Inferring Ancestor-Descendant Relationships in the Fossil Record (With Statistics) David Bapst, Melanie Hopkins, April Wright, Nick Matzke & Graeme Lloyd GSA 2016 T151 Wednesday Sept 28 th, 9:15 AM Feel
17.2 Classification Based on Evolutionary Relationships Organization of all that speciation!
 Organization of all that speciation! Patterns of evolution.. Taxonomy gets an over haul! Using more than morphology! 3 domains, 6 kingdoms KEY CONCEPT Modern classification is based on evolutionary relationships.
Organization of all that speciation! Patterns of evolution.. Taxonomy gets an over haul! Using more than morphology! 3 domains, 6 kingdoms KEY CONCEPT Modern classification is based on evolutionary relationships.
Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per.
 Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per. Introduction Imagine a single diagram representing the evolutionary relationships between everything that has ever lived. If life evolved
Interpreting Evolutionary Trees Honors Integrated Science 4 Name Per. Introduction Imagine a single diagram representing the evolutionary relationships between everything that has ever lived. If life evolved
Chapter 13. Phylogenetic Systematics: Developing an Hypothesis of Amniote Relationships
 Chapter 3 Phylogenetic Systematics: Developing an Hypothesis of Amniote Relationships Daniel R. Brooks, Deborah A. McLennan, Joseph P. Carney Michael D. Dennison, and Corey A. Goldman Department of Zoology
Chapter 3 Phylogenetic Systematics: Developing an Hypothesis of Amniote Relationships Daniel R. Brooks, Deborah A. McLennan, Joseph P. Carney Michael D. Dennison, and Corey A. Goldman Department of Zoology
Are node-based and stem-based clades equivalent? Insights from graph theory
 Are node-based and stem-based clades equivalent? Insights from graph theory November 18, 2010 Tree of Life 1 2 Jeremy Martin, David Blackburn, E. O. Wiley 1 Associate Professor of Mathematics, San Francisco,
Are node-based and stem-based clades equivalent? Insights from graph theory November 18, 2010 Tree of Life 1 2 Jeremy Martin, David Blackburn, E. O. Wiley 1 Associate Professor of Mathematics, San Francisco,
Understanding Evolutionary History: An Introduction to Tree Thinking
 1 Understanding Evolutionary History: An Introduction to Tree Thinking Laura R. Novick Kefyn M. Catley Emily G. Schreiber Vanderbilt University Western Carolina University Vanderbilt University Version
1 Understanding Evolutionary History: An Introduction to Tree Thinking Laura R. Novick Kefyn M. Catley Emily G. Schreiber Vanderbilt University Western Carolina University Vanderbilt University Version
Do the traits of organisms provide evidence for evolution?
 PhyloStrat Tutorial Do the traits of organisms provide evidence for evolution? Consider two hypotheses about where Earth s organisms came from. The first hypothesis is from John Ray, an influential British
PhyloStrat Tutorial Do the traits of organisms provide evidence for evolution? Consider two hypotheses about where Earth s organisms came from. The first hypothesis is from John Ray, an influential British
Ch 1.2 Determining How Species Are Related.notebook February 06, 2018
 Name 3 "Big Ideas" from our last notebook lecture: * * * 1 WDYR? Of the following organisms, which is the closest relative of the "Snowy Owl" (Bubo scandiacus)? a) barn owl (Tyto alba) b) saw whet owl
Name 3 "Big Ideas" from our last notebook lecture: * * * 1 WDYR? Of the following organisms, which is the closest relative of the "Snowy Owl" (Bubo scandiacus)? a) barn owl (Tyto alba) b) saw whet owl
DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS
 Syst. Biol. 45(4):39^14, 1996 DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS STEVEN POE Department of Zoology and Texas Memorial Museum, University of Texas, Austin, Texas 78712-1064, USA; E-mail:
Syst. Biol. 45(4):39^14, 1996 DATA SET INCONGRUENCE AND THE PHYLOGENY OF CROCODILIANS STEVEN POE Department of Zoology and Texas Memorial Museum, University of Texas, Austin, Texas 78712-1064, USA; E-mail:
Required and Recommended Supporting Information for IUCN Red List Assessments
 Required and Recommended Supporting Information for IUCN Red List Assessments This is Annex 1 of the Rules of Procedure for IUCN Red List Assessments 2017 2020 as approved by the IUCN SSC Steering Committee
Required and Recommended Supporting Information for IUCN Red List Assessments This is Annex 1 of the Rules of Procedure for IUCN Red List Assessments 2017 2020 as approved by the IUCN SSC Steering Committee
Unveiling Vicariant Methodologies in Vicariance Biogeography NOT ANYTHING GOES. Marco G.P. van Veller
 Unveiling Vicariant Methodologies in Vicariance Biogeography NOT ANYTHING GOES Marco G.P. van Veller Illustratie omslag: Rita van Veller-Keizer Veller, Marco G.P. van Unveiling vicariant methodologies
Unveiling Vicariant Methodologies in Vicariance Biogeography NOT ANYTHING GOES Marco G.P. van Veller Illustratie omslag: Rita van Veller-Keizer Veller, Marco G.P. van Unveiling vicariant methodologies
The impact of the recognizing evolution on systematics
 The impact of the recognizing evolution on systematics 1. Genealogical relationships between species could serve as the basis for taxonomy 2. Two sources of similarity: (a) similarity from descent (b)
The impact of the recognizing evolution on systematics 1. Genealogical relationships between species could serve as the basis for taxonomy 2. Two sources of similarity: (a) similarity from descent (b)
LABORATORY #10 -- BIOL 111 Taxonomy, Phylogeny & Diversity
 LABORATORY #10 -- BIOL 111 Taxonomy, Phylogeny & Diversity Scientific Names ( Taxonomy ) Most organisms have familiar names, such as the red maple or the brown-headed cowbird. However, these familiar names
LABORATORY #10 -- BIOL 111 Taxonomy, Phylogeny & Diversity Scientific Names ( Taxonomy ) Most organisms have familiar names, such as the red maple or the brown-headed cowbird. However, these familiar names
Phylogenetics. Phylogenetic Trees. 1. Represent presumed patterns. 2. Analogous to family trees.
 Phylogenetics. Phylogenetic Trees. 1. Represent presumed patterns of descent. 2. Analogous to family trees. 3. Resolve taxa, e.g., species, into clades each of which includes an ancestral taxon and all
Phylogenetics. Phylogenetic Trees. 1. Represent presumed patterns of descent. 2. Analogous to family trees. 3. Resolve taxa, e.g., species, into clades each of which includes an ancestral taxon and all
Taxonomy and Pylogenetics
 Taxonomy and Pylogenetics Taxonomy - Biological Classification First invented in 1700 s by Carolus Linneaus for organizing plant and animal species. Based on overall anatomical similarity. Similarity due
Taxonomy and Pylogenetics Taxonomy - Biological Classification First invented in 1700 s by Carolus Linneaus for organizing plant and animal species. Based on overall anatomical similarity. Similarity due
Darwin s Finches: A Thirty Year Study.
 Darwin s Finches: A Thirty Year Study. I. Mit-DNA Based Phylogeny (Figure 1). 1. All Darwin s finches descended from South American grassquit (small finch) ancestor circa 3 Mya. 2. Galapagos colonized
Darwin s Finches: A Thirty Year Study. I. Mit-DNA Based Phylogeny (Figure 1). 1. All Darwin s finches descended from South American grassquit (small finch) ancestor circa 3 Mya. 2. Galapagos colonized
These small issues are easily addressed by small changes in wording, and should in no way delay publication of this first- rate paper.
 Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper reports on a highly significant discovery and associated analysis that are likely to be of broad interest to the scientific community.
Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper reports on a highly significant discovery and associated analysis that are likely to be of broad interest to the scientific community.
May 10, SWBAT analyze and evaluate the scientific evidence provided by the fossil record.
 May 10, 2017 Aims: SWBAT analyze and evaluate the scientific evidence provided by the fossil record. Agenda 1. Do Now 2. Class Notes 3. Guided Practice 4. Independent Practice 5. Practicing our AIMS: E.3-Examining
May 10, 2017 Aims: SWBAT analyze and evaluate the scientific evidence provided by the fossil record. Agenda 1. Do Now 2. Class Notes 3. Guided Practice 4. Independent Practice 5. Practicing our AIMS: E.3-Examining
Testing Phylogenetic Hypotheses with Molecular Data 1
 Testing Phylogenetic Hypotheses with Molecular Data 1 How does an evolutionary biologist quantify the timing and pathways for diversification (speciation)? If we observe diversification today, the processes
Testing Phylogenetic Hypotheses with Molecular Data 1 How does an evolutionary biologist quantify the timing and pathways for diversification (speciation)? If we observe diversification today, the processes
Comparing DNA Sequences Cladogram Practice
 Name Period Assignment # See lecture questions 75, 122-123, 127, 137 Comparing DNA Sequences Cladogram Practice BACKGROUND Between 1990 2003, scientists working on an international research project known
Name Period Assignment # See lecture questions 75, 122-123, 127, 137 Comparing DNA Sequences Cladogram Practice BACKGROUND Between 1990 2003, scientists working on an international research project known
TOPIC CLADISTICS
 TOPIC 5.4 - CLADISTICS 5.4 A Clades & Cladograms https://upload.wikimedia.org/wikipedia/commons/thumb/4/46/clade-grade_ii.svg IB BIO 5.4 3 U1: A clade is a group of organisms that have evolved from a common
TOPIC 5.4 - CLADISTICS 5.4 A Clades & Cladograms https://upload.wikimedia.org/wikipedia/commons/thumb/4/46/clade-grade_ii.svg IB BIO 5.4 3 U1: A clade is a group of organisms that have evolved from a common
Guide to the Professional Practice Standard: Veterinarian-Client-Patient Relationship (VCPR)
 Guide to the Professional Practice Standard: Veterinarian-Client-Patient Relationship (VCPR) Published October 2018 This College publication describes a mandatory standard of practice. The Veterinarians
Guide to the Professional Practice Standard: Veterinarian-Client-Patient Relationship (VCPR) Published October 2018 This College publication describes a mandatory standard of practice. The Veterinarians
The Origin of Species: Lizards in an Evolutionary Tree
 The Origin of Species: Lizards in an Evolutionary Tree NAME DATE This handout supplements the short film The Origin of Species: Lizards in an Evolutionary Tree. 1. Puerto Rico, Cuba, Jamaica, and Hispaniola
The Origin of Species: Lizards in an Evolutionary Tree NAME DATE This handout supplements the short film The Origin of Species: Lizards in an Evolutionary Tree. 1. Puerto Rico, Cuba, Jamaica, and Hispaniola
6. The lifetime Darwinian fitness of one organism is greater than that of another organism if: A. it lives longer than the other B. it is able to outc
 1. The money in the kingdom of Florin consists of bills with the value written on the front, and pictures of members of the royal family on the back. To test the hypothesis that all of the Florinese $5
1. The money in the kingdom of Florin consists of bills with the value written on the front, and pictures of members of the royal family on the back. To test the hypothesis that all of the Florinese $5
Evolution in Action: Graphing and Statistics
 Evolution in Action: Graphing and Statistics OVERVIEW This activity serves as a supplement to the film The Origin of Species: The Beak of the Finch and provides students with the opportunity to develop
Evolution in Action: Graphing and Statistics OVERVIEW This activity serves as a supplement to the film The Origin of Species: The Beak of the Finch and provides students with the opportunity to develop
Postilla PEABODY MUSEUM OF NATURAL HISTORY YALE UNIVERSITY NEW HAVEN, CONNECTICUT, U.S.A.
 Postilla PEABODY MUSEUM OF NATURAL HISTORY YALE UNIVERSITY NEW HAVEN, CONNECTICUT, U.S.A. Number 117 18 March 1968 A 7DIAPSID (REPTILIA) PARIETAL FROM THE LOWER PERMIAN OF OKLAHOMA ROBERT L. CARROLL REDPATH
Postilla PEABODY MUSEUM OF NATURAL HISTORY YALE UNIVERSITY NEW HAVEN, CONNECTICUT, U.S.A. Number 117 18 March 1968 A 7DIAPSID (REPTILIA) PARIETAL FROM THE LOWER PERMIAN OF OKLAHOMA ROBERT L. CARROLL REDPATH
The Making of the Fittest: LESSON STUDENT MATERIALS USING DNA TO EXPLORE LIZARD PHYLOGENY
 The Making of the Fittest: Natural The The Making Origin Selection of the of Species and Fittest: Adaptation Natural Lizards Selection in an Evolutionary and Adaptation Tree INTRODUCTION USING DNA TO EXPLORE
The Making of the Fittest: Natural The The Making Origin Selection of the of Species and Fittest: Adaptation Natural Lizards Selection in an Evolutionary and Adaptation Tree INTRODUCTION USING DNA TO EXPLORE
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution
 Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
Bioinformatics: Investigating Molecular/Biochemical Evidence for Evolution Background How does an evolutionary biologist decide how closely related two different species are? The simplest way is to compare
PROBLEMS DUE TO MISSING DATA IN PHYLOGENETIC ANALYSES INCLUDING FOSSILS: A CRITICAL REVIEW
 Journal of Vertebrate Paleontology 23(2):263-274, June 2003 0 2003 by the Society of Vertebrate Paleontology 1, PROLMS U TO MISSING T IN PHYLOGNTI NLYSS INLUING FOSSILS: RITIL RVIW MURN KRNY and JMS M.
Journal of Vertebrate Paleontology 23(2):263-274, June 2003 0 2003 by the Society of Vertebrate Paleontology 1, PROLMS U TO MISSING T IN PHYLOGNTI NLYSS INLUING FOSSILS: RITIL RVIW MURN KRNY and JMS M.
8/19/2013. What is convergence? Topic 11: Convergence. What is convergence? What is convergence? What is convergence? What is convergence?
 Topic 11: Convergence What are the classic herp examples? Have they been formally studied? Emerald Tree Boas and Green Tree Pythons show a remarkable level of convergence Photos KP Bergmann, Philadelphia
Topic 11: Convergence What are the classic herp examples? Have they been formally studied? Emerald Tree Boas and Green Tree Pythons show a remarkable level of convergence Photos KP Bergmann, Philadelphia
Comparing DNA Sequence to Understand
 Comparing DNA Sequence to Understand Evolutionary Relationships with BLAST Name: Big Idea 1: Evolution Pre-Reading In order to understand the purposes and learning objectives of this investigation, you
Comparing DNA Sequence to Understand Evolutionary Relationships with BLAST Name: Big Idea 1: Evolution Pre-Reading In order to understand the purposes and learning objectives of this investigation, you
Sample Questions: EXAMINATION I Form A Mammalogy -EEOB 625. Name Composite of previous Examinations
 Sample Questions: EXAMINATION I Form A Mammalogy -EEOB 625 Name Composite of previous Examinations Part I. Define or describe only 5 of the following 6 words - 15 points (3 each). If you define all 6,
Sample Questions: EXAMINATION I Form A Mammalogy -EEOB 625 Name Composite of previous Examinations Part I. Define or describe only 5 of the following 6 words - 15 points (3 each). If you define all 6,
1 Describe the anatomy and function of the turtle shell. 2 Describe respiration in turtles. How does the shell affect respiration?
 GVZ 2017 Practice Questions Set 1 Test 3 1 Describe the anatomy and function of the turtle shell. 2 Describe respiration in turtles. How does the shell affect respiration? 3 According to the most recent
GVZ 2017 Practice Questions Set 1 Test 3 1 Describe the anatomy and function of the turtle shell. 2 Describe respiration in turtles. How does the shell affect respiration? 3 According to the most recent
Evolution. Evolution is change in organisms over time. Evolution does not have a goal; it is often shaped by natural selection (see below).
 Evolution Evolution is change in organisms over time. Evolution does not have a goal; it is often shaped by natural selection (see below). Species an interbreeding population of organisms that can produce
Evolution Evolution is change in organisms over time. Evolution does not have a goal; it is often shaped by natural selection (see below). Species an interbreeding population of organisms that can produce
Chapter 16: Evolution Lizard Evolution Virtual Lab Honors Biology. Name: Block: Introduction
 Chapter 16: Evolution Lizard Evolution Virtual Lab Honors Biology Name: Block: Introduction Charles Darwin proposed that over many generations some members of a population could adapt to a changing environment
Chapter 16: Evolution Lizard Evolution Virtual Lab Honors Biology Name: Block: Introduction Charles Darwin proposed that over many generations some members of a population could adapt to a changing environment
Evolution and Biodiversity Laboratory Systematics and Taxonomy I. Taxonomy taxonomy taxa taxon taxonomist natural artificial systematics
 Evolution and Biodiversity Laboratory Systematics and Taxonomy by Dana Krempels and Julian Lee Recent estimates of our planet's biological diversity suggest that the species number between 5 and 50 million,
Evolution and Biodiversity Laboratory Systematics and Taxonomy by Dana Krempels and Julian Lee Recent estimates of our planet's biological diversity suggest that the species number between 5 and 50 million,
Warm-Up: Fill in the Blank
 Warm-Up: Fill in the Blank 1. For natural selection to happen, there must be variation in the population. 2. The preserved remains of organisms, called provides evidence for evolution. 3. By using and
Warm-Up: Fill in the Blank 1. For natural selection to happen, there must be variation in the population. 2. The preserved remains of organisms, called provides evidence for evolution. 3. By using and
PHYLOGENETIC TAXONOMY*
 Annu. Rev. Ecol. Syst. 1992.23:449~0 PHYLOGENETIC TAXONOMY* Kevin dd Queiroz Division of Amphibians and Reptiles, United States National Museum of Natural History, Smithsonian Institution, Washington,
Annu. Rev. Ecol. Syst. 1992.23:449~0 PHYLOGENETIC TAXONOMY* Kevin dd Queiroz Division of Amphibians and Reptiles, United States National Museum of Natural History, Smithsonian Institution, Washington,
Biology 2108 Laboratory Exercises: Variation in Natural Systems. LABORATORY 2 Evolution: Genetic Variation within Species
 Biology 2108 Laboratory Exercises: Variation in Natural Systems Ed Bostick Don Davis Marcus C. Davis Joe Dirnberger Bill Ensign Ben Golden Lynelle Golden Paula Jackson Ron Matson R.C. Paul Pam Rhyne Gail
Biology 2108 Laboratory Exercises: Variation in Natural Systems Ed Bostick Don Davis Marcus C. Davis Joe Dirnberger Bill Ensign Ben Golden Lynelle Golden Paula Jackson Ron Matson R.C. Paul Pam Rhyne Gail
1.3. Initial training shall include sufficient obedience training to perform an effective and controlled search.
 SWGDOG SC 9 - HUMAN SCENT DOGS Scent Identification Lineups Posted for Public Comment 9/2/2008 11/1/2008. Posted for Public Comment 1/19/2010 3/19/2010. Approved by the membership 3/3/2010. Scent identification
SWGDOG SC 9 - HUMAN SCENT DOGS Scent Identification Lineups Posted for Public Comment 9/2/2008 11/1/2008. Posted for Public Comment 1/19/2010 3/19/2010. Approved by the membership 3/3/2010. Scent identification
Modern taxonomy. Building family trees 10/10/2011. Knowing a lot about lots of creatures. Tom Hartman. Systematics includes: 1.
 Modern taxonomy Building family trees Tom Hartman www.tuatara9.co.uk Classification has moved away from the simple grouping of organisms according to their similarities (phenetics) and has become the study
Modern taxonomy Building family trees Tom Hartman www.tuatara9.co.uk Classification has moved away from the simple grouping of organisms according to their similarities (phenetics) and has become the study
Clinical trials conducted in subjects with naturally
 Review J Vet Intern Med 2013 Evidence-Based Medicine: The Design and Interpretation of Noninferiority Clinical Trials in Veterinary Medicine K.J. Freise, T.-L. Lin, T.M. Fan, V. Recta, and T.P. Clark Noninferiority
Review J Vet Intern Med 2013 Evidence-Based Medicine: The Design and Interpretation of Noninferiority Clinical Trials in Veterinary Medicine K.J. Freise, T.-L. Lin, T.M. Fan, V. Recta, and T.P. Clark Noninferiority
Pedigrees: Understanding Retriever Pedigrees Part I
 Pedigrees: Understanding Retriever Pedigrees Part I Written by Butch Goodwin of Northern Flight Retrievers Editor's Note -Reading and understanding pedigrees is vital to picking out a sound, healthy puppy.
Pedigrees: Understanding Retriever Pedigrees Part I Written by Butch Goodwin of Northern Flight Retrievers Editor's Note -Reading and understanding pedigrees is vital to picking out a sound, healthy puppy.
Biodiversity and Distributions. Lecture 2: Biodiversity. The process of natural selection
 Lecture 2: Biodiversity What is biological diversity? Natural selection Adaptive radiations and convergent evolution Biogeography Biodiversity and Distributions Types of biological diversity: Genetic diversity
Lecture 2: Biodiversity What is biological diversity? Natural selection Adaptive radiations and convergent evolution Biogeography Biodiversity and Distributions Types of biological diversity: Genetic diversity
Evolution as Fact. The figure below shows transitional fossils in the whale lineage.
 Evolution as Fact Evolution is a fact. Organisms descend from others with modification. Phylogeny, the lineage of ancestors and descendants, is the scientific term to Darwin's phrase "descent with modification."
Evolution as Fact Evolution is a fact. Organisms descend from others with modification. Phylogeny, the lineage of ancestors and descendants, is the scientific term to Darwin's phrase "descent with modification."
The Origin of Species: Lizards in an Evolutionary Tree
 The Origin of Species: Lizards in an Evolutionary Tree Cara Larracas, Stacy Lopez, Takara Yaegashi Period 4 Background Information Throughout the Caribbean Islands there is a species of anole lizards that
The Origin of Species: Lizards in an Evolutionary Tree Cara Larracas, Stacy Lopez, Takara Yaegashi Period 4 Background Information Throughout the Caribbean Islands there is a species of anole lizards that
EVOLUTIONARY GENETICS (Genome 453) Midterm Exam Name KEY
 PLEASE: Put your name on every page and SHOW YOUR WORK. Also, lots of space is provided, but you do not have to fill it all! Note that the details of these problems are fictional, for exam purposes only.
PLEASE: Put your name on every page and SHOW YOUR WORK. Also, lots of space is provided, but you do not have to fill it all! Note that the details of these problems are fictional, for exam purposes only.
Clarifications to the genetic differentiation of German Shepherds
 Clarifications to the genetic differentiation of German Shepherds Our short research report on the genetic differentiation of different breeding lines in German Shepherds has stimulated a lot interest
Clarifications to the genetic differentiation of German Shepherds Our short research report on the genetic differentiation of different breeding lines in German Shepherds has stimulated a lot interest
Evolution of Birds. Summary:
 Oregon State Standards OR Science 7.1, 7.2, 7.3, 7.3S.1, 7.3S.2 8.1, 8.2, 8.2L.1, 8.3, 8.3S.1, 8.3S.2 H.1, H.2, H.2L.4, H.2L.5, H.3, H.3S.1, H.3S.2, H.3S.3 Summary: Students create phylogenetic trees to
Oregon State Standards OR Science 7.1, 7.2, 7.3, 7.3S.1, 7.3S.2 8.1, 8.2, 8.2L.1, 8.3, 8.3S.1, 8.3S.2 H.1, H.2, H.2L.4, H.2L.5, H.3, H.3S.1, H.3S.2, H.3S.3 Summary: Students create phylogenetic trees to
Indigo Sapphire Bear. Newfoundland. Indigo Sapphire Bear. January. Dog's name: DR. NEALE FRETWELL. R&D Director
 Indigo Sapphire Bear Dog's name: Indigo Sapphire Bear This certifies the authenticity of Indigo Sapphire Bear's canine genetic background as determined following careful analysis of more than 300 genetic
Indigo Sapphire Bear Dog's name: Indigo Sapphire Bear This certifies the authenticity of Indigo Sapphire Bear's canine genetic background as determined following careful analysis of more than 300 genetic
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST In this laboratory investigation, you will use BLAST to compare several genes, and then use the information to construct a cladogram.
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST In this laboratory investigation, you will use BLAST to compare several genes, and then use the information to construct a cladogram.
Original language: English PC22 Doc. 10 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA
 Original language: English PC22 Doc. 10 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA Twenty-second meeting of the Plants Committee Tbilisi (Georgia), 19-23 October 2015
Original language: English PC22 Doc. 10 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA Twenty-second meeting of the Plants Committee Tbilisi (Georgia), 19-23 October 2015
8/19/2013. Topic 5: The Origin of Amniotes. What are some stem Amniotes? What are some stem Amniotes? The Amniotic Egg. What is an Amniote?
 Topic 5: The Origin of Amniotes Where do amniotes fall out on the vertebrate phylogeny? What are some stem Amniotes? What is an Amniote? What changes were involved with the transition to dry habitats?
Topic 5: The Origin of Amniotes Where do amniotes fall out on the vertebrate phylogeny? What are some stem Amniotes? What is an Amniote? What changes were involved with the transition to dry habitats?
Darwin and the Family Tree of Animals
 Darwin and the Family Tree of Animals Note: These links do not work. Use the links within the outline to access the images in the popup windows. This text is the same as the scrolling text in the popup
Darwin and the Family Tree of Animals Note: These links do not work. Use the links within the outline to access the images in the popup windows. This text is the same as the scrolling text in the popup
Answers to Questions about Smarter Balanced 2017 Test Results. March 27, 2018
 Answers to Questions about Smarter Balanced Test Results March 27, 2018 Smarter Balanced Assessment Consortium, 2018 Table of Contents Table of Contents...1 Background...2 Jurisdictions included in Studies...2
Answers to Questions about Smarter Balanced Test Results March 27, 2018 Smarter Balanced Assessment Consortium, 2018 Table of Contents Table of Contents...1 Background...2 Jurisdictions included in Studies...2
STATUS SIGNALING IN DARK-EYED JUNCOS
 STATUS SIGNALING IN DARK-EYED JUNCOS ELLEN D. KETTERSON Department of Biology, Indiana University, Bloomington, Indiana 47401 USA ABSTR CT.--Rohwer (1975, 1977) has proposed that members of certain variably-plumaged
STATUS SIGNALING IN DARK-EYED JUNCOS ELLEN D. KETTERSON Department of Biology, Indiana University, Bloomington, Indiana 47401 USA ABSTR CT.--Rohwer (1975, 1977) has proposed that members of certain variably-plumaged
Systematics and taxonomy of the genus Culicoides what is coming next?
 Systematics and taxonomy of the genus Culicoides what is coming next? Claire Garros 1, Bruno Mathieu 2, Thomas Balenghien 1, Jean-Claude Delécolle 2 1 CIRAD, Montpellier, France 2 IPPTS, Strasbourg, France
Systematics and taxonomy of the genus Culicoides what is coming next? Claire Garros 1, Bruno Mathieu 2, Thomas Balenghien 1, Jean-Claude Delécolle 2 1 CIRAD, Montpellier, France 2 IPPTS, Strasbourg, France
Identity Management with Petname Systems. Md. Sadek Ferdous 28th May, 2009
 Identity Management with Petname Systems Md. Sadek Ferdous 28th May, 2009 Overview Entity, Identity, Identity Management History and Rationales Components and Properties Application Domain of Petname Systems
Identity Management with Petname Systems Md. Sadek Ferdous 28th May, 2009 Overview Entity, Identity, Identity Management History and Rationales Components and Properties Application Domain of Petname Systems
Let s Build a Cladogram!
 Name Let s Build a Cladogram! Date Introduction: Cladistics is one of the newest trends in the modern classification of organisms. This method shows the relationship between different organisms based on
Name Let s Build a Cladogram! Date Introduction: Cladistics is one of the newest trends in the modern classification of organisms. This method shows the relationship between different organisms based on
Who Cares? The Evolution of Parental Care in Squamate Reptiles. Ben Halliwell Geoffrey While, Tobias Uller
 Who Cares? The Evolution of Parental Care in Squamate Reptiles Ben Halliwell Geoffrey While, Tobias Uller 1 Parental Care any instance of parental investment that increases the fitness of offspring 2 Parental
Who Cares? The Evolution of Parental Care in Squamate Reptiles Ben Halliwell Geoffrey While, Tobias Uller 1 Parental Care any instance of parental investment that increases the fitness of offspring 2 Parental
American Rescue Dog Association. Standards and Certification Procedures
 American Rescue Dog Association Standards and Certification Procedures American Rescue Dog Association Section II Area Search Certification Date Last Updated: October 2014 Date Last Reviewed: May 2016
American Rescue Dog Association Standards and Certification Procedures American Rescue Dog Association Section II Area Search Certification Date Last Updated: October 2014 Date Last Reviewed: May 2016
The Accuracy of M ethods for C oding and Sampling Higher-Lev el Tax a for Phylogenetic Analysis: A Simulatio n Study
 Syst. Biol. 47(3): 397 ± 413, 1998 The Accuracy of M ethods for C oding and Sampling Higher-Lev el Tax a for Phylogenetic Analysis: A Simulatio n Study JO HN J. W IENS Section of Amphibians and Reptiles,
Syst. Biol. 47(3): 397 ± 413, 1998 The Accuracy of M ethods for C oding and Sampling Higher-Lev el Tax a for Phylogenetic Analysis: A Simulatio n Study JO HN J. W IENS Section of Amphibians and Reptiles,
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Types of Evolution: Punctuated Equilibrium vs Gradualism
 Biology Types of Evolution: Punctuated Equilibrium vs Gradualism Use the information below AND YOUR NOTES to answer the questions that follow. READ the information before attempting to do the work. You
Biology Types of Evolution: Punctuated Equilibrium vs Gradualism Use the information below AND YOUR NOTES to answer the questions that follow. READ the information before attempting to do the work. You
SHEEP SIRE REFERENCING SCHEMES - NEW OPPORTUNITIES FOR PEDIGREE BREEDERS AND LAMB PRODUCERS a. G. Simm and N.R. Wray
 SHEEP SIRE REFERENCING SCHEMES - NEW OPPORTUNITIES FOR PEDIGREE BREEDERS AND LAMB PRODUCERS a G. Simm and N.R. Wray The Scottish Agricultural College Edinburgh, Scotland Summary Sire referencing schemes
SHEEP SIRE REFERENCING SCHEMES - NEW OPPORTUNITIES FOR PEDIGREE BREEDERS AND LAMB PRODUCERS a G. Simm and N.R. Wray The Scottish Agricultural College Edinburgh, Scotland Summary Sire referencing schemes
Name Class Date. How does a founding population adapt to new environmental conditions?
 Open-Ended Inquiry Skills Lab Additional Lab 8 Ecosystems and Speciation Problem How does a founding population adapt to new environmental conditions? Introduction When the hurricane s winds died down,
Open-Ended Inquiry Skills Lab Additional Lab 8 Ecosystems and Speciation Problem How does a founding population adapt to new environmental conditions? Introduction When the hurricane s winds died down,
The Divergence of the Marine Iguana: Amblyrhyncus cristatus. from its earlier land ancestor (what is now the Land Iguana). While both the land and
 Chris Lang Course Paper Sophomore College October 9, 2008 Abstract--- The Divergence of the Marine Iguana: Amblyrhyncus cristatus In this course paper, I address the divergence of the Galapagos Marine
Chris Lang Course Paper Sophomore College October 9, 2008 Abstract--- The Divergence of the Marine Iguana: Amblyrhyncus cristatus In this course paper, I address the divergence of the Galapagos Marine
Title. Grade level. Time. Student Target. PART 3 Lesson: Populations. PART 3 Activity: Turtles, Turtle Everywhere! minutes
 Title PART 3 Lesson: Populations PART 3 Activity: Turtles, Turtle Everywhere! Grade level 3-5 Time 60 minutes Student Target SC.3.N.1.1 Raise questions about the natural world, investigate them individually
Title PART 3 Lesson: Populations PART 3 Activity: Turtles, Turtle Everywhere! Grade level 3-5 Time 60 minutes Student Target SC.3.N.1.1 Raise questions about the natural world, investigate them individually
Phylogeny of Harpacticoida (Copepoda): Revision of Maxillipedasphalea and Exanechentera
 Phylogeny of Harpacticoida (Copepoda): Revision of Maxillipedasphalea and Exanechentera Sybille Seifried sybille.seifried@mail.uni-oldenburg.de published 2003 by Cuvillier Verlag, Göttingen ISBN 3-89873-845-0
Phylogeny of Harpacticoida (Copepoda): Revision of Maxillipedasphalea and Exanechentera Sybille Seifried sybille.seifried@mail.uni-oldenburg.de published 2003 by Cuvillier Verlag, Göttingen ISBN 3-89873-845-0
Phylogeny of genus Vipio latrielle (Hymenoptera: Braconidae) and the placement of Moneilemae group of Vipio species based on character weighting
 International Journal of Biosciences IJB ISSN: 2220-6655 (Print) 2222-5234 (Online) http://www.innspub.net Vol. 3, No. 3, p. 115-120, 2013 RESEARCH PAPER OPEN ACCESS Phylogeny of genus Vipio latrielle
International Journal of Biosciences IJB ISSN: 2220-6655 (Print) 2222-5234 (Online) http://www.innspub.net Vol. 3, No. 3, p. 115-120, 2013 RESEARCH PAPER OPEN ACCESS Phylogeny of genus Vipio latrielle
Mendelian Genetics Using Drosophila melanogaster Biology 12, Investigation 1
 Mendelian Genetics Using Drosophila melanogaster Biology 12, Investigation 1 Learning the rules of inheritance is at the core of all biologists training. These rules allow geneticists to predict the patterns
Mendelian Genetics Using Drosophila melanogaster Biology 12, Investigation 1 Learning the rules of inheritance is at the core of all biologists training. These rules allow geneticists to predict the patterns
HONR219D Due 3/29/16 Homework VI
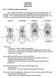 Part 1: Yet More Vertebrate Anatomy!!! HONR219D Due 3/29/16 Homework VI Part 1 builds on homework V by examining the skull in even greater detail. We start with the some of the important bones (thankfully
Part 1: Yet More Vertebrate Anatomy!!! HONR219D Due 3/29/16 Homework VI Part 1 builds on homework V by examining the skull in even greater detail. We start with the some of the important bones (thankfully
& chicken. Antibiotic Resistance
 Antibiotic Resistance & chicken Chicken Farmers of Canada (CFC) supports the judicious use of antibiotics that have been approved by the Veterinary Drugs Directorate of Health Canada, in order to ensure
Antibiotic Resistance & chicken Chicken Farmers of Canada (CFC) supports the judicious use of antibiotics that have been approved by the Veterinary Drugs Directorate of Health Canada, in order to ensure
BioSci 110, Fall 08 Exam 2
 1. is the cell division process that results in the production of a. mitosis; 2 gametes b. meiosis; 2 gametes c. meiosis; 2 somatic (body) cells d. mitosis; 4 somatic (body) cells e. *meiosis; 4 gametes
1. is the cell division process that results in the production of a. mitosis; 2 gametes b. meiosis; 2 gametes c. meiosis; 2 somatic (body) cells d. mitosis; 4 somatic (body) cells e. *meiosis; 4 gametes
Natural Selection and the Evolution of Darwin s Finches. Activity Student Handout
 Natural Selection and the Evolution of Darwin s Finches INTRODUCTION There are 13 different species of finch on the Galápagos Islands off the coast of Ecuador. On one of the islands, Daphne Major, biologists
Natural Selection and the Evolution of Darwin s Finches INTRODUCTION There are 13 different species of finch on the Galápagos Islands off the coast of Ecuador. On one of the islands, Daphne Major, biologists
2013 AVMA Veterinary Workforce Summit. Workforce Research Plan Details
 2013 AVMA Veterinary Workforce Summit Workforce Research Plan Details If the American Veterinary Medical Association (AVMA) says the profession is experiencing a 12.5 percent excess capacity in veterinary
2013 AVMA Veterinary Workforce Summit Workforce Research Plan Details If the American Veterinary Medical Association (AVMA) says the profession is experiencing a 12.5 percent excess capacity in veterinary
INHERITANCE OF BODY WEIGHT IN DOMESTIC FOWL. Single Comb White Leghorn breeds of fowl and in their hybrids.
 440 GENETICS: N. F. WATERS PROC. N. A. S. and genetical behavior of this form is not incompatible with the segmental interchange theory of circle formation in Oenothera. Summary.-It is impossible for the
440 GENETICS: N. F. WATERS PROC. N. A. S. and genetical behavior of this form is not incompatible with the segmental interchange theory of circle formation in Oenothera. Summary.-It is impossible for the
